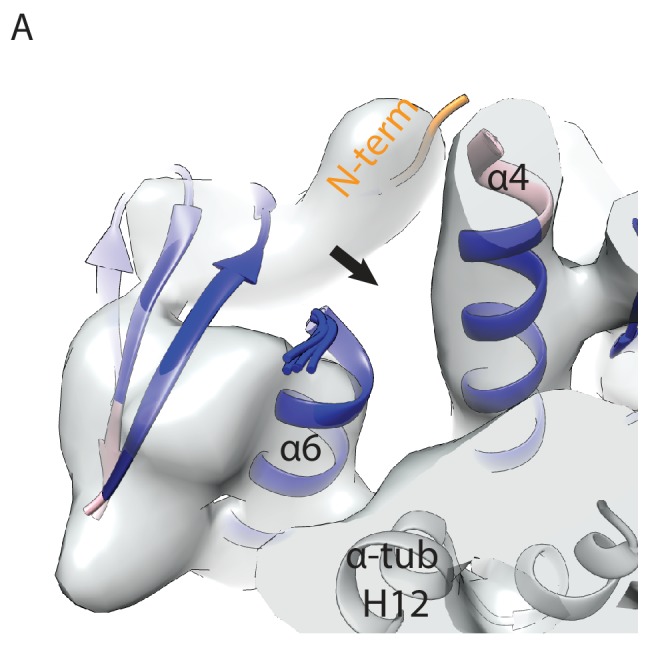
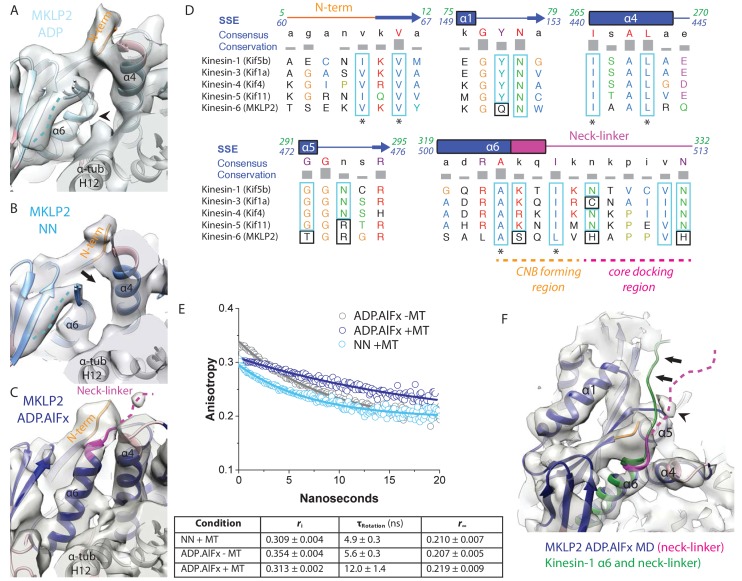
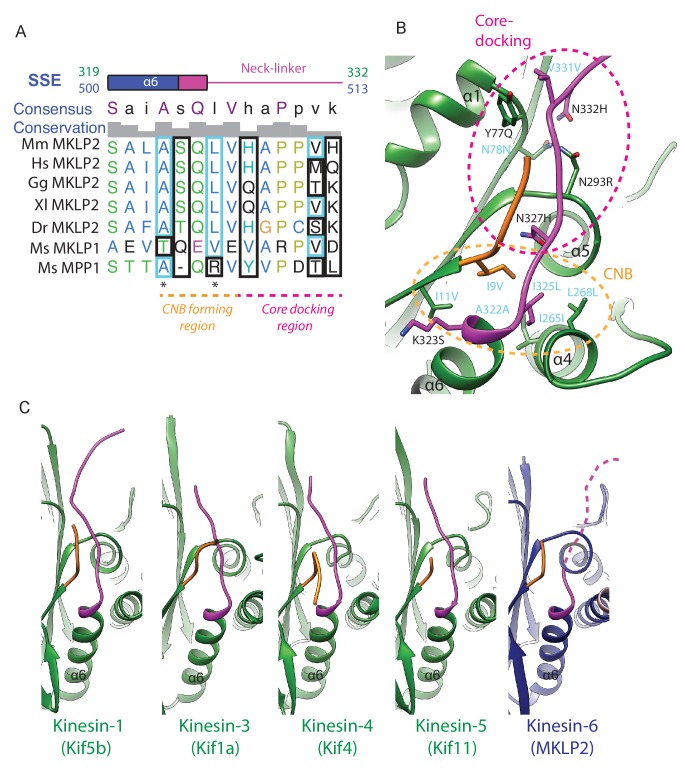
Figure 6. MKLP2 neck-linker response to nucleotide.
(A) MKLP2-MD-ADP has a short helix-α6 leaving a gap between its terminus and helix-α4 (arrow), showing the cleft for the neck linker is closed and the CNB is not formed. Helix-α6 has relatively poor density, suggesting it may be partially disordered at its C-terminus (arrowhead). Density that could flexibly accommodate part of the neck linker is present connected to β-sheet1 (blue dotted line). (B) MKLP2-MD-NN also has a short helix-α6 leaving a gap between its terminus and helix-α4 (arrow). (C) MKLP2-MD-ADP.AlFx has an extended helix-α6, the initial portion of the neck-linker (magenta) inserts between the N-terminus (orange) and helix-α4 forming the CNB (arrowhead). The remaining portion of the neck-linker is flexible. (D) Sequence alignments of representative members (in brackets) of the Kin1/3/4/5/6 families for the neck-linker and its contact regions. Residue colouring using the Clustal X scheme (Larkin et al., 2007). Sequence numberings for Kif5b (Kin1, green) and MKLP2 (kinesin-6, blue) are shown adjacent to the secondary structure schematics. Well conserved residues in MKLP2 are boxed in light blue, whereas otherwise well conserved residues which have diverged in MKLP2 are boxed in black (see also Figure 6—figure supplement 2). Asterisks indicate conserved hydrophobic residues participating in CNB formation. Regions of the neck-linker involved in CNB formation or core docking are indicated by dashed lines in orange and magenta, respectively. (E) Time-resolved fluorescence anisotropy of FlAsH labeled MKLP2-MD: (1) NN + MT, light blue; (2) ADP.AlFx –MT, grey and (3) ADP.AlFx +MT, dark blue. Data shown in the table below are representative of 5 replicate samples. (F) MKLP2-MD-ADP.AlFx neck-linker in magenta (disordered region, dashed line). Kin1 helix-α6 and neck-linker (dark green) has been superimposed on helix-α6 of MKLP2-MD, showing the expected position of the neck-linker. There is no density corresponding to a docked neck-linker (arrows) suggesting it is mainly disordered. A small amount of density close to helix-α5 (arrowhead) likely indicates that alternative conformations are flexibly explored.
Figure 6—figure supplement 1. The neck-linker in MT-bound MKLP2-MD-AMPPNP is not directed towards the MT plus end.