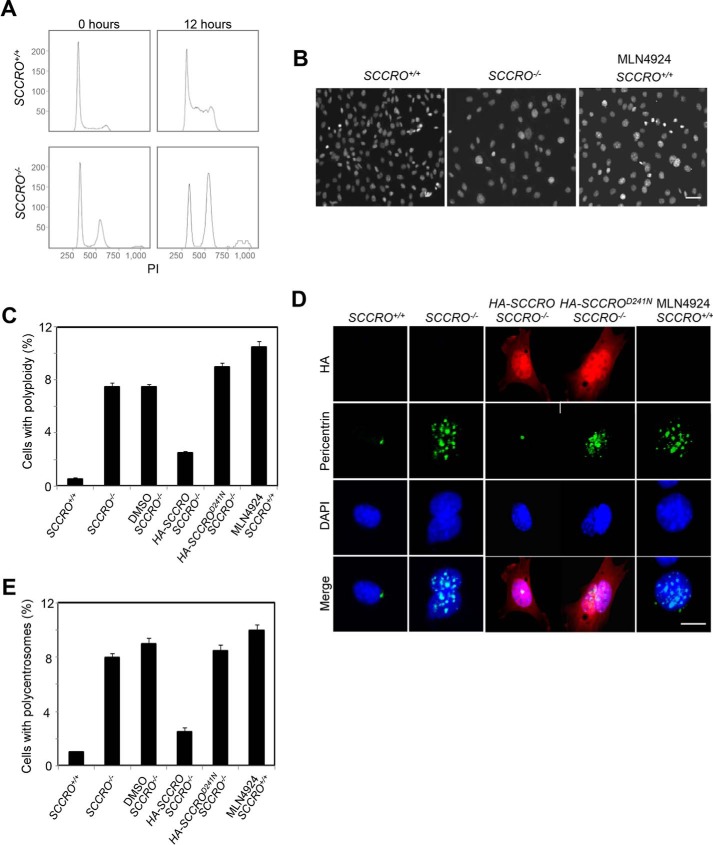
Figure 1.
Targeted disruption of SCCRO results in a defect in mitosis. A, flow cytometry analysis (propidium iodide (PI) staining) on MEFs released from synchronization by serum starvation, showing polyploid accumulated in SCCRO−/− MEFs. B, DAPI staining for DNA content confirmed polyploid accumulated in SCCRO−/− MEFs and MLN4924-treated (0.5 μm for 12 h) SCCRO+/+ MEFs. A representative result from staining of SCCRO+/+ MEFs shows an absence of polyploidy. Scale bar = 20 μm. C, quantification of polyploidy in SCCRO+/+ and SCCRO−/− MEFs, SCCRO−/− MEFs infected with a retrovirus carrying SCCRO or SCCROD241N cDNA, and SCCRO+/+ MEFs treated with MLN4924. Polyploidy in SCCRO−/− MEFs could be rescued by overexpression of HA-SCCRO but not the neddylation-dead mutant HA-SCCROD241N. D, immunofluorescence using α-pericentrin antibody, showing normal centrosome numbers and localization in SCCRO+/+ but malpositioned and supernumerary centrosomes in SCCRO−/− MEFs and MLN4924-treated SCCRO+/+ MEFs. The centrosome defect in SCCRO−/− MEFs could be rescued by overexpression of HA-SCCRO but not HA-SCCROD241N. Scale bar = 5 μm. E, quantification of the percentage of polycentrosome cells for C.