Figure 6.
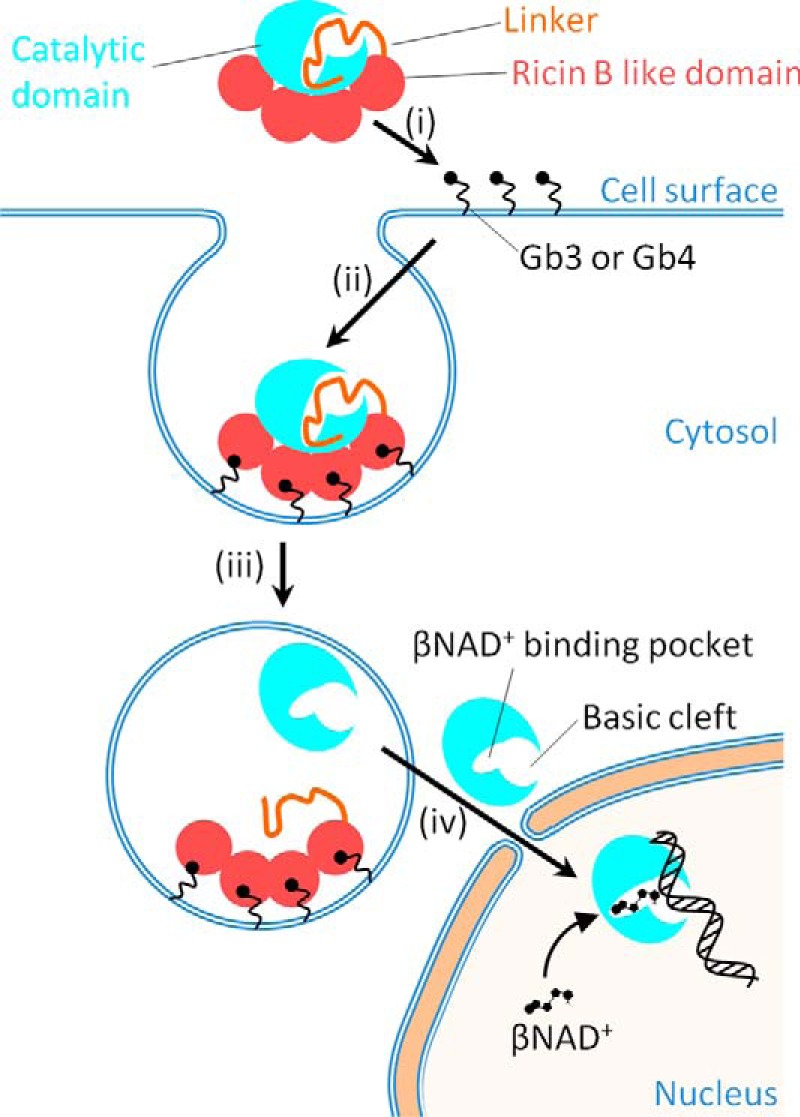
A model for the activation and DNA binding of DNA ADP-ribosylating pierisin-1. Pierisin-1 consists of an N-terminal catalytic domain colored light blue, a linker shown as an orange thread, and a C-terminal ricin B–like domain colored red and autoinhibits a DNA ADP-ribosylating activity by occupying its catalytic cleft with its linker region. i and ii, pierisin binds to cell surface glycosphingolipid receptors, such as globotriaosylceramide (Gb3) and globotetraosylceramide (Gb4) through its four ricin B–like domains (13) and is internalized into cells. iii, the pierisin incorporated into lysozomes is cleaved at a C-terminal site of the catalytic domain and then followed by the catalytic domain dissociation from the rest of the linker and the ricin B–like domains, resulting in the activation of its catalytic domain. Thus, the catalytic domain is exposed at a catalytic site and freely accessible by a substrate βNAD+ and an acceptor DNA. iv, the active catalytic domain (residues 1–233) released into the cytosol migrates into the nucleus and binds to DNA, where it transfers the ADP-ribose moiety of βNAD+ to the N2 amino group of guanine.
