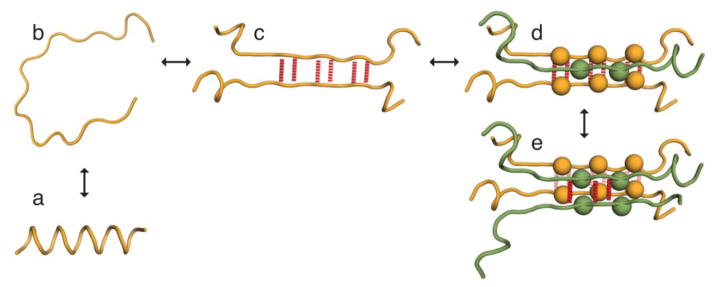
Figure 1.
Cartoon of the aggregation states modeled in our theory. The left panels show the conversion of monomers between (a) an aggregation-resistant folded state that we model as a helix and (b) an aggregation-prone disordered state. (c) A pair of (partially) unfolded proteins forming a dimer through the formation of intermolecular H-bonds. In the figure the dimer is held together by m2 = 6 H-bonds (red dotted lines). (d) A trimer which is held together by H-bonds between the first two molecules (rear) and steric zipper interactions with the third molecule (front). Sidechains participating in the steric zipper are shown as spheres with the remaining sidechains omitted for clarity. The total number of ordered amino acids is 2m2 + m3, m2 from each of the rear molecules plus m3 from the front molecule. In the illustrated conformation there are two sidechains from the front molecule participating in the steric zipper. Since every other sidechain is on the opposite face of the β-strand, this requires at least m3 = 3 amino acids to be in the β-sheet conformation. (e) A tetramer with m2 = 6 β-ordered amino acids on each of the molecules in the rear β-sheet (six yellow H-bonds) and three β ordered amino acids on each strand of the front β-sheet (three red H-bonds).