Fig. 1.
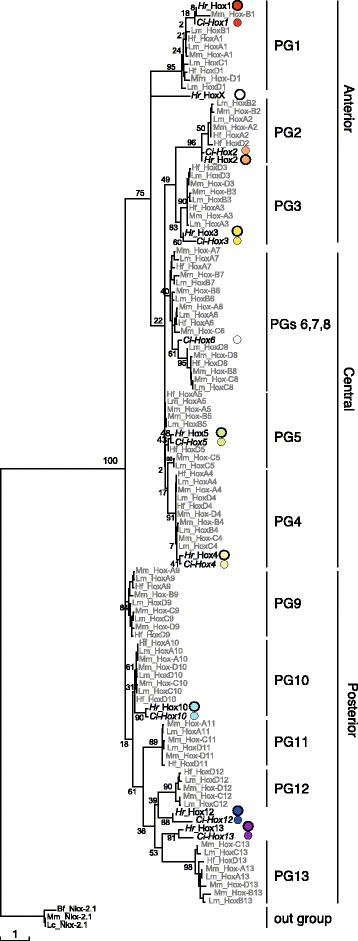
Phylogenetic analysis of Hox gene candidates of Halocynthia roretzi. The ML tree was constructed using homeodomain sequences and the adjacent 20 N-terminal and seven C-terminal amino acid residues (Additional file 1: Figure S1) and MEGA5 software package. The percentage of 1000 replicated trees in which clustering of genes was supported is indicated at nodes. Within a clade consisting only of vertebrate Hox genes, the percentage is not indicated. Hr Hox gene candidates and Hox genes of Ciona intestinalis (Ci-Hox) are indicated by larger and smaller colored circles, respectively. Here, Hr Hox gene candidates are tentatively designated according to their Ci counterparts, except for HoxX, which did not show apparent orthology to Ci-Hox genes. The color-code indicates distinct paralog groups (PGs). Taxonomic abbreviations are Mm for Mus musculus, Lm for Latimeria menadoensis, Hf for Heterodontus francisci, Ci for Ciona intestinalis and Hr for Halocynthia roretzi. The bar at the bottom indicates one amino acid substitution per position in the sequence
