Fig. 4.
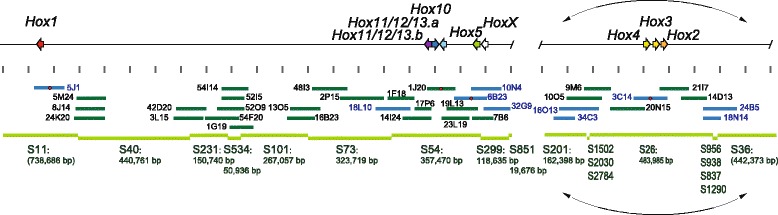
Schematic representation of the Hox gene cluster of Halocynthia roretzi. The Hox gene cluster structure of Hr as estimated by chromosome/scaffold walking is shown at the top. A horizontal line represents a part of chromosome. The telomeric side is to the left and the centromeric side is to the right. Hox genes are represented as thick arrows, which also indicate transcription direction. The color code is the same as that in Fig. 1. Between HoxX and Hox4, there is a region, from which no clones were available out of the BAC library; hence, no scaffolds available out of the ANISEED database. Grey arrays of short vertical bars indicate 100 kbp, starting at the right and left ends of the region where no scaffolds are available. Dark green or blue horizontal bars below the scales indicate BAC clones corresponding to Hox gene clusters. Dark green bars indicate BAC clones, end regions of which were sequenced. Blue bars indicate BAC clones for which the whole insert sequence was determined. Bars with red dots in the middle are the clones used for probes for chromosomal FISH (Fig. 3). Names of clones are also indicated. Green horizontal bars at the bottom indicate scaffolds in the ANISEED database (Halocynthia roretzi MTP2014; https://www.aniseed.cnrs.fr/fgb2/gbrowse/harore_mtp2014/) that correspond to the Hox gene cluster spanning chromosomal region. The length of each scaffold according to ANISEED database is indicated below scaffold names, except for the scaffolds in the two regions adjacent to S26, where multiple small scaffolds are included. Arc lines with arrowheads on both ends placed upper and lower of the subcluster region of Hox2, 3 and 4 indicate that the orientation of the subcluster has not been determined
