Fig. 2.
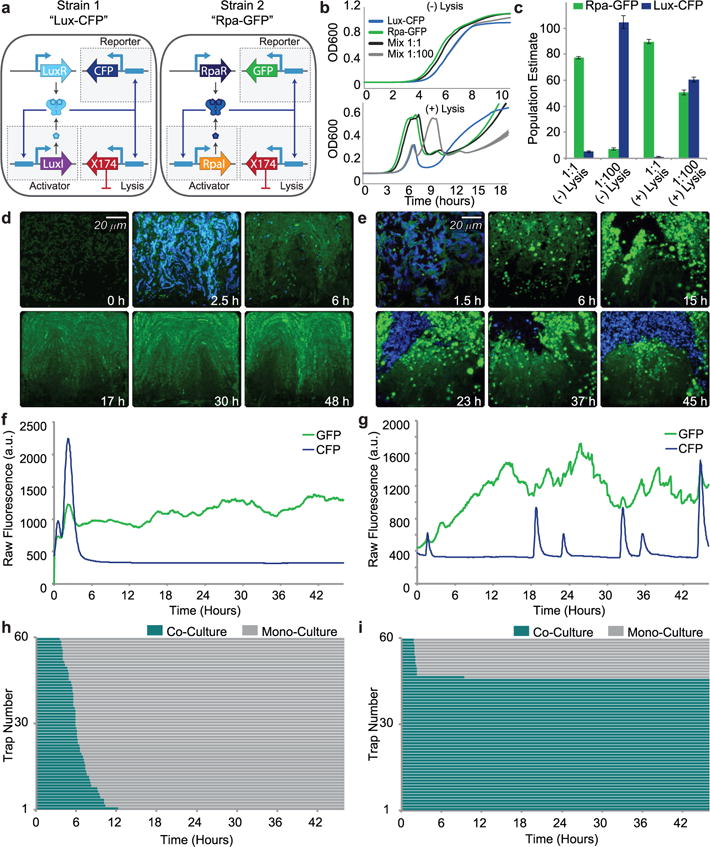
Experimental demonstration of long-term co-culture of competitive species with unequal growth rates using signal orthogonal self-lysis a, Genetic diagram of a two-strain ecosystem of self-lysing Salmonella constructed with two signal orthogonal quorum sensing systems, rpa and lux. b, Batch culture growth curves of the Lux-CFP strain alone (blue), Rpa-GFP strain alone (green), a 1:1 mixture (black), and a 1:100 (Rpa-GFP:Lux-CFP) mixture (gray), both without the lysis gene (top) and with the lysis gene (bottom). All strains were started from the same diluted density and under the same growth conditions. Width of lines represent s.d. (n=3) c, Batch culture population estimates of Lux-CFP and Rpa-GFP co-cultures. Rpa-GFP population estimated as GFP fluorescence (integrated over the full length of the experiment) of the mixture normalized by the time-integrated GFP fluorescence of Rpa-GFP cells alone. Lux-CFP population estimated as CFP fluorescence (integrated over the full length of the experiment) of the mixture normalized by the time-integrated CFP fluorescence Lux-CFP cells alone. Error bars represent s.d. (n=3) d, Video stills of a representative co-culture of non-lysing Lux-CFP and Rpa-GFP strains showing the eventual takeover by the green strain. e, Video stills of a representative co-culture of the Lux-CFP and Rpa-GFP strains with the lysis plasmid. The addition of the lysis plasmid prevents either strain from taking over for the duration of the experiment. f, Time trace of the GFP and CFP Fluorescence of the trap in the video shown in d. g, Time trace of the GFP and CFP Fluorescence of the trap in the video shown in e. h, Graph showing the length of co-culture for each of the sixty traps containing the non-lysing strains. i, Graph showing the length of co-culture for each of the sixty traps containing the strains with the lysis plasmids.
