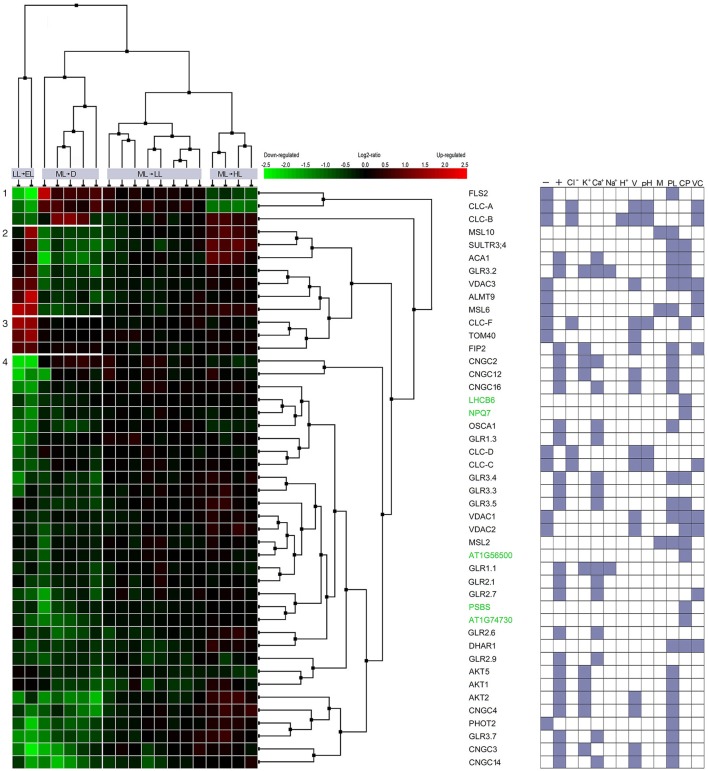
Figure 3.
Co-expression of light-responsive genes involved in the flow of ions across the membranes and in the non-photochemical quenching. Multiple microarray and RNAseq experiments, as well as NCBI database were searched to select genes which respond to the excess light (induction of SAA and SAR) and are involved in ion transport across biological membranes in Arabidopsis. 273 genes with query “ion channel” and 10 genes for query “nonphotochemical quenching” were verified in the AmiGO 2 database. Finally, the Similarity Search Tool and Hierarchical Clustering of the Genevestigator were used to select 45 genes with the most closely correlated expression profiles during defined light conditions (Exp ID: AT-00467 and AT-00682). The transfer of the plant from low light (LL, 65 μmol photons m−2 s−1) to excess light (EL, 1300 μmol photons m−2 s−1), from moderate light (ML, 150 μmol photons m−2 s−1) to darkness (D), from ML to LL; and from ML to high light (HL, 400 μmol photons m−2 s−1) was compared. Left panel represents relative expression data of experimental vs control values. GO class was assigned to each gene according to NCBI and Gene Set Enrichment tool of Genevestigetor and the results is indicated in the right panel as: - anion channel activity; + cation channel activity; Cl−, K+, Ca2+ Na+, H+, chloride, potassium, calcium, sodium and hydrogen ions channel activity; V, pH, M - voltage-, pH-, mechanically-gated ion channel activity; PL, CP, VC, protein localization in plasma membrane, chloroplast, vacuole.