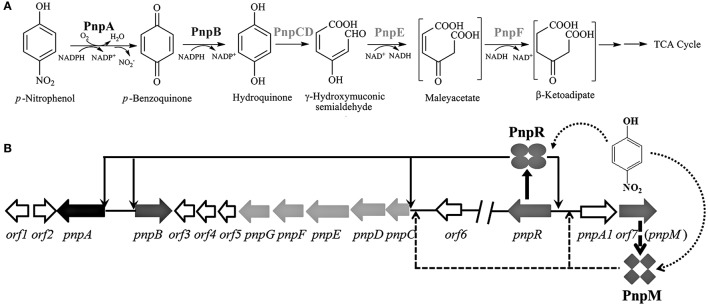
Figure 1.
(A) Proposed pathway for PNP catabolism in Pseudomonas sp. strain WBC-3 (modified after Zhang et al., 2009) pnpA and pnpB are PNP 4-monooxygenase and p-benzoquinone reductase respectively; pnpCDEF encode α- and β-subunits of hydroquinone dioxygenase, γ-hydroxymuconic semialdehyde dehydrogenase and maleylacetate reductase, respectively. (B) Schematic of the regulatory circuit of the PNP catabolic cluster in Pseudomonas sp. strain WBC-3 (figure not drawn to scale, modified after Zhang et al., 2015). Four ellipses stand for PnpR tetramer, and four diamonds stand for PnpM tetramer. The curved and dotted arrow stands for binding of para-nitrophenol (PNP) with PnpR or PnpM. The down arrows stand for PnpR's activating four operons of pnpA, pnpB, pnpCDEFG, and pnpR. The up arrows stand for PnpM's activating two operons of pnpCDEFG and pnpA1M.PnpR regulates transcription initiation in four regions pnpA, pnpB, pnpCDEFG, and pnpR. PnpM regulates transcription initiation in two regions pnpCDEFG and pnpA1M.