Figure 1.
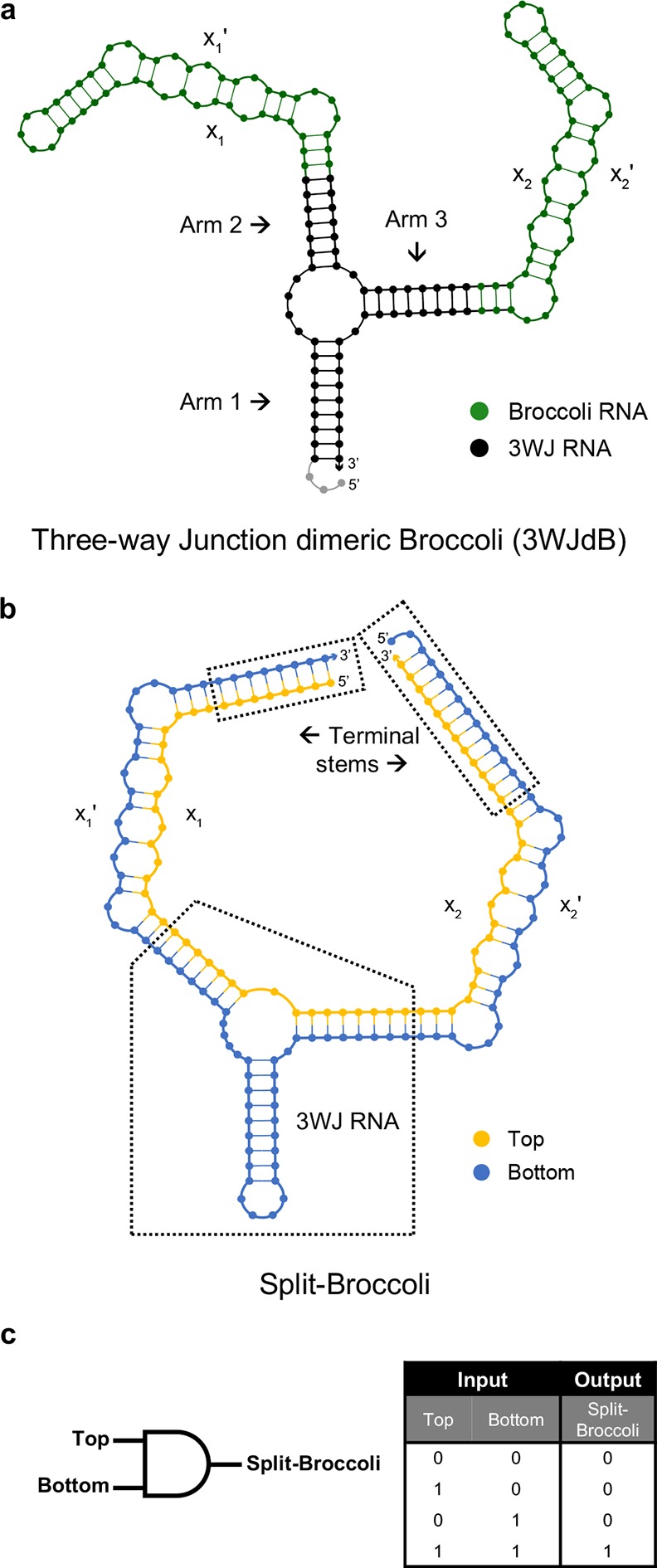
Design and NUPACK predicted secondary structures of 3WJdB and Split-Broccoli. (a) Monomers of Broccoli aptamer (green) were inserted into Arms 2 and 3 of the three-way junction (3WJ) RNA motif (black) to create the unimolecular, unsplit three-way junction dimeric Broccoli (3WJdB). (b) Design of the Split-Broccoli system required inversion of the Broccoli monomer present in Arm 2 to ensure that neither Top (yellow) nor Bottom (blue) strand alone contained the full sequence required to independently form a fully functional monomer of Broccoli (i.e., xn:xn′). Predicted hybridization of the two strands was strengthened with the addition of terminal stems. (c) The Split-Broccoli system illustrated as an RNA AND gate (left) and its corresponding truth table (right). Output from the system should be true (1) only when both inputs are also true.
