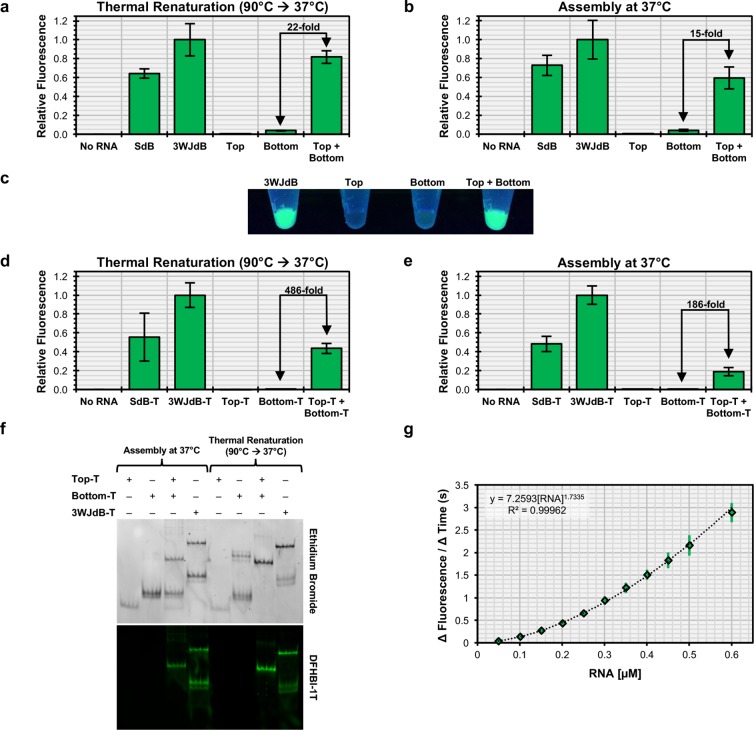
Figure 2.
In vitro assembly of the Split-Broccoli system in the absence and presence of transcription terminator structures. Assembly of equimolar amounts of purified Split-Broccoli RNA components (Top + Bottom) demonstrate robust function comparable to fluorescence of the stabilized dimeric Broccoli (SdB) and 3WJdB, (a) when thermally renatured or (b) when simply incubated together at physiological temperature. Background signal from either Top or Bottom alone remains minimal for both assembly methods. (c) Fluorescence of 3WJdB and the Split-System (Top + Bottom, thermally renatured) is easily observed when excited with longwave ultraviolet light, whereas signal from Bottom alone is only slightly discernible. When transcribed with transcription terminator structures (denoted by appending “-T” to the names of the individual RNAs) and assembled in vitro (d, e), the Split-Broccoli system exhibits a decrease in relative fluorescence, but demonstrates a larger fold-change in fluorescence activation over either Top-T or Bottom-T alone. (f) Nondenaturing gel electrophoresis and dual staining with ethidium bromide and DFHBI-1T of the Split-Broccoli system with transcription terminator structures suggests that decreased fluorescence of the system is a result of incomplete hybridization between Top-T and Bottom-T, rather than nonfunctional assembly. (g) Functional assembly of Top-T and Bottom-T approximately follows second-order kinetics (y = A[Top][Bottom] = A[Top]2, for equimolar mixture). Mean values are shown with error bars to indicate standard deviations (n = 5 for panels a, b, d, e; n = 4 for panel g).