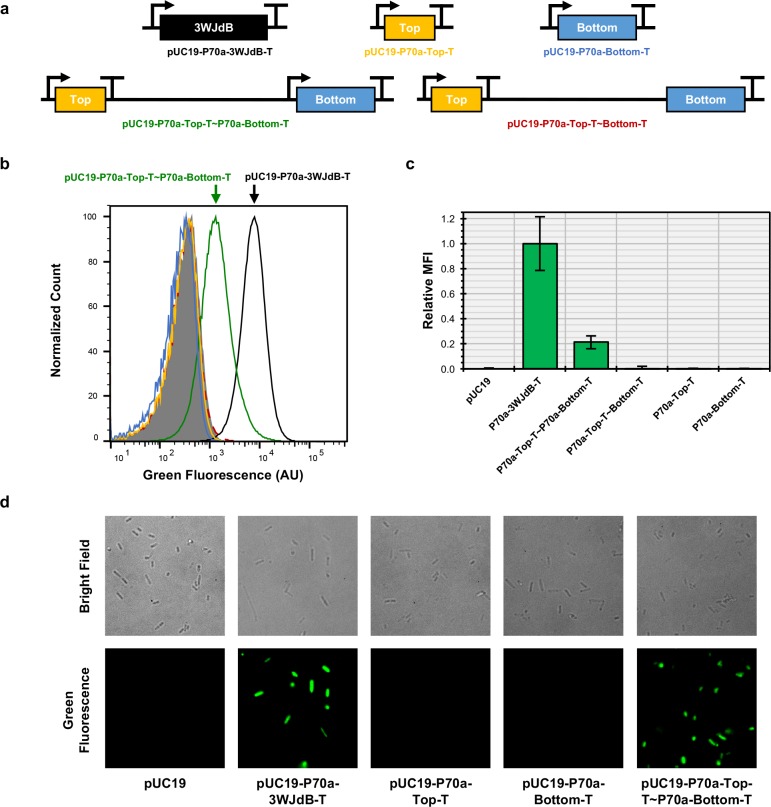
Figure 4.
The Split-Broccoli system functions when expressed in vivo. (a) DNA templates corresponding to 3WJdB-T (black), Top-T (yellow), Bottom-T (blue) were individually cloned into the pUC19 plasmid. A single plasmid expressing both Top-T and Bottom-T was created (pUC19-P70a-Top-T∼P70a-Bottom-T), as was a control plasmid for runon transcription which lacked a promoter immediately upstream of Bottom-T (pUC19-P70a-Top-T∼Bottom-T). (b) A representative flow cytometry histogram of 5 × 104 events per population illustrates a shift in fluorescence for the plasmid containing the Split-Broccoli expression plasmid (green). Bacterial populations transformed with plasmids containing either Top-T or Bottom-T alone, or lacking a promoter upstream of Bottom-T, demonstrate background levels of fluorescence equivalent to the unmodified pUC19 plasmid control. (c) Relative mean fluorescence intensities for flow cytometric analyses of transformed populations, normalized to the pUC19 plasmid (set to 0) and 3WJdB-T (set to 1), are shown with error bars to indicate standard deviations (n ≥ 4). (d) Fluorescence microscopy imaging further validates the in vivo functionality of the Split-Broccoli system, as green fluorescence is only observed for E. coli transformed with either the unimolecular 3WJdB-T encoding plasmid or bimolecular Split-Broccoli encoding plasmid.