Figure 1. Effect of MUC16 Expression on SKOV3 and A2780 Ovarian Cancer Cells.
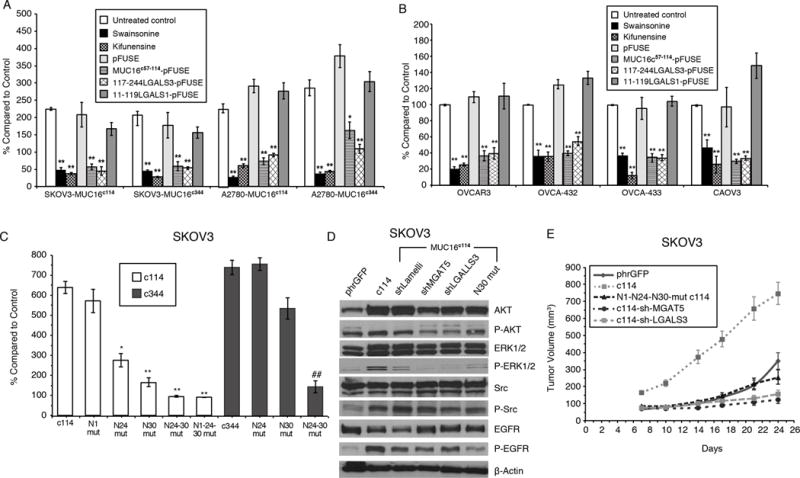
A) MUC16 enhancement of matrigel invasion assay SKOV3 and A2780 cells depends on N-glycosylation. Cells were exposed to Swainsonine(1 ug/ml), Kifunensine (1ug/ml), control pFUSE protein, MUC16c57-114-pFUSE fusion protein, 117-244LGALS3-pFUSE, or 11-119LGALS1-pFUSE fusion protein (all 5 ug/ml) and compared to vector controls. Results (n=3) are expressed as percentages compared by paired t test to parent SKOV3 orA2780 phrGFP control after 48 h (Mean ± SE). (*=p,0.01; ** p< 0.0001) In both SKOV3 and A2780 cell lines, MUC16c114 or MUC16c344 cells were more invasive than the control phrGFP cells (** = p<0.0001), and these invasive properties were unaffected by exposure to the pFUSE vector-only protein. Each study was replicated ≥ 3 times.
B) Matrigel invasion assay for wild-type ovarian cancer cell lines depends on Galectin-3. OVCAR3, OVCA-432, OVCA-433, and CAOV3 cells were exposed to Swainsosine (1 ug/ml), Kifunensin (1ug/m), control pFUSE protein, MUC16c57-114-pFUSE fusion protein, 117-244LGALS3-pFUSE, or 11-119LGALS1-pFUSE fusion protein (all 5 ug/ml). Invasion was measured in triplicate (n=3) and normalized against untreated control cells. Results (n=3) are expressed as percentages compared by paired t test to parental control cells after 48 h (Mean ± SE). (*=p,0.01; ** p< 0.0001) Two or more replicates were performed for each condition.
C) Loss of proximal N-glycosylation sites impair matrigel invasion. SKOV3-MUC16c114 and SKOV3-MUC16c344 transfected cell lines were tested for MUC16-based increased invasion following N→A mutations of N-glycosylation sites at each of the N1, N24, and N30 positions. Invasion was measured in triplicate (n=3), with 3 or more independent replicates. Results are expressed as percentages compared to SKOV3phr cells without MUC16 expression. Results (n=3) are expressed as percentages compared by paired t test to parental control cells after 48 h (Mean ± SE). (*=p,0.01; ** p< 0.0001)
D) MUC16-induced oncogene activation on AKT, MAPK, and SRC signaling pathways indicating MUC16 increased phosphorylation of AKT (S473), ERK1/2 (pT202/Y204), SRC(y416), and EGFR(pY1068) in SKOV3-MUC16c114 cells. MGAT5 (shMGAT5), Galectin-3 (shLGALS3) knockdowns, and N30A mutation all reduced MUC16c114-induced oncogene activation.
E) MUC16 N-glycosylation–dependent tumor growth in vivo. In vivo growth of SKOV3-MUC16c114 expressing cell line was much more aggressive (p<0.0001 by paired t test) compared to the SKOV3-phrGFP control. However, SKOV3-MUC16(N1-N24-N30)mut-c114 glycosylation -impaired transfectants did not show any growth enhancement compared to SKOV3-phrGFP vector control tumors. SKOV-3-MUC16c114 cells with shRNA of LGALS3 or MGAT5 were also similar to control cells
