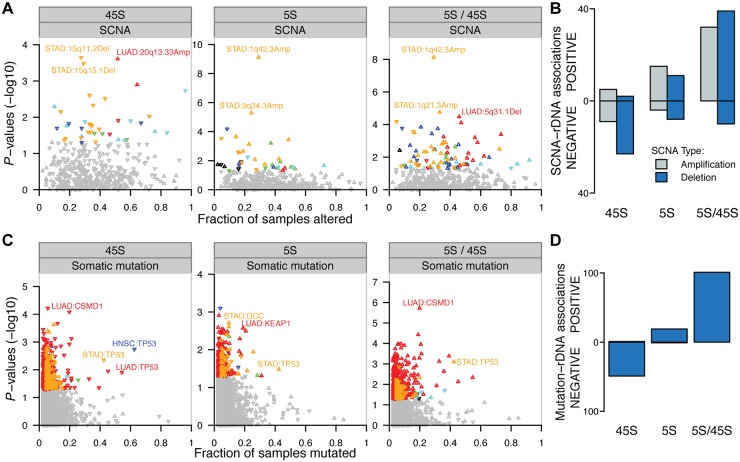
Fig 4. Association between genetic context and rDNA CN alterations.
(A) Associations between copy number alterations (SCNA) and 5S CN, 45S CN and 5S/45S ratio. (B) Significantly associated rDNA-SCNA pairs (P < 0.05) are preferentially implicated in greater 45S loss and greater 5S rDNA amplification (P < 0.05, binomial test). (C) Association between somatic mutations and 5S CN, 45S CN and 5S/45S ratio. (D) Significantly associated mutation-rDNA pairs (P < 0.05) are almost exclusively implicated in greater 45S loss and greater 5S rDNA amplification (P < 0.001, binomial test) in LUAD. For (B, C) Y-axis show the P-values for the associations between the SCNA or gene mutation event and 45S CN, 5S CN and 5S/45S ratio. rDNA associations were colored according to cancer type (P < 0.05). The up/down direction of triangles indicates that the somatic alteration is associated with increased or decreased CN or 5S/45S ratio. The X-axis shows the fraction of patients with the non-silent gene mutation or focal SCNAs (ploidy cutoff > 2.1 for amplification and < 1.9 for deletion).