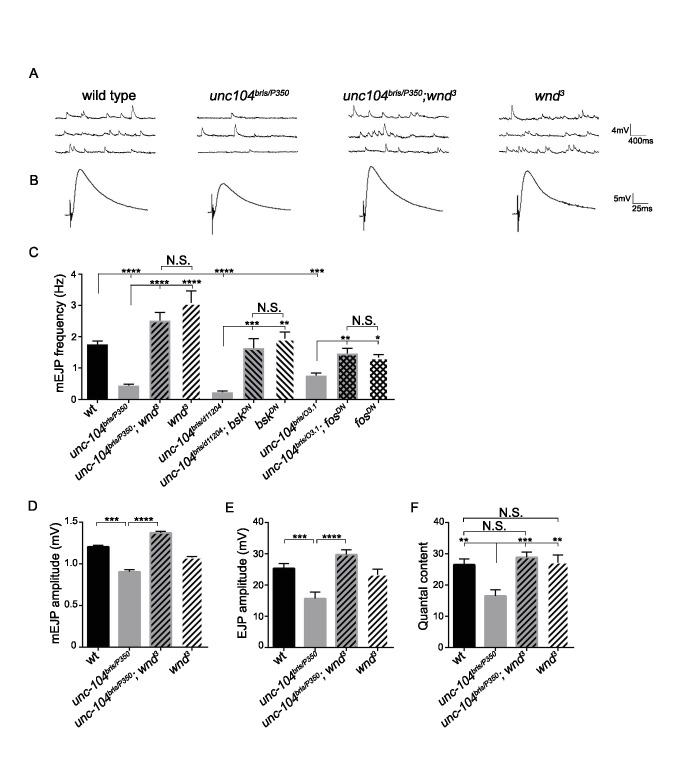
Figure 3. The synaptic transmission defect in unc-104 mutants is suppressed by wnd mutations.
(A–B) Representative electrophysiological traces of (A) miniature Excitatory Junctional Potentials (mEJP) and (B) Evoked Excitatory Junctional Potentials (EJP) recorded from muscle 6 of third instar larvae. (C) Quantification of impaired mEJP frequency for unc-104-hypomorph mutants and their rescue by wnd mutations or expression of dominant negative (DN) bsk or fos (driven by elav-Gal4 and BG380-Gal4, respectively). Representative traces for bskDN and fosDN are shown in Figure 3—figure supplement 1. (D–F) Quantification of (D) mEJP amplitude, (E) EJP amplitude and (F) quantal content (corrected for nonlinear summation). See also Figure 3—figure supplement 1 for additional quantification of bskDN and fosDN data. Unc-10403.1 and unc-104bris are hypomorphic alleles, while unc-104P350 and unc-104d11204 are null alleles. wnd3 is a presumptive null mutation in wnd (Collins et al., 2006). wt animals are Canton S. All data are represented as mean ±SEM; N.S., not significant; ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05. Tukey test for multiple comparison. For additional data, see Figure 3—figure supplements 1–4.
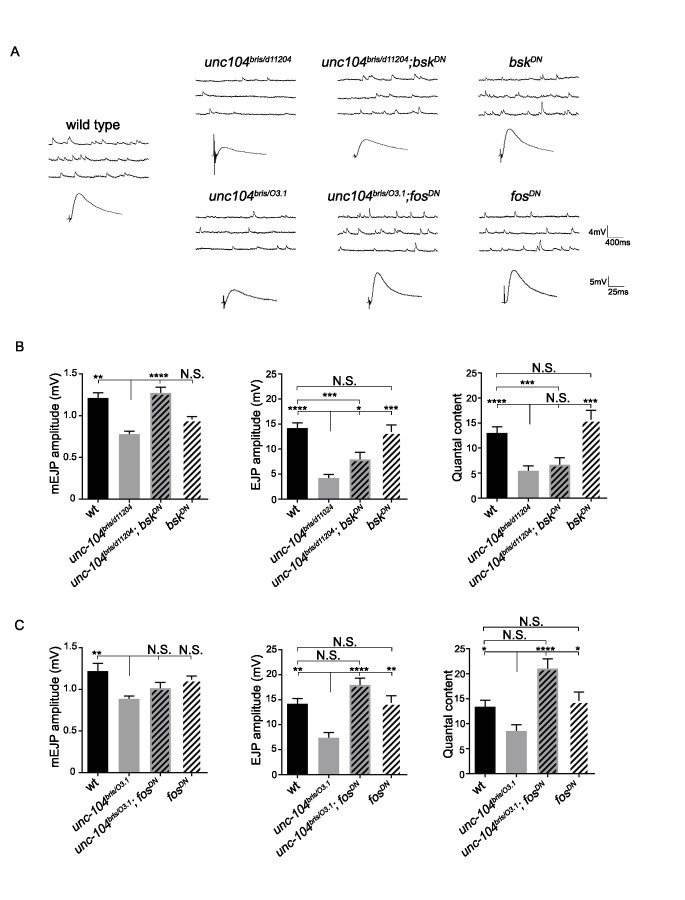
Figure 3—figure supplement 1. Wnd signaling components JNK/Bsk and Fos promotes synaptic transmission defects in unc-104-hypomorph mutants, (related to Figure 3).
Figure 3—figure supplement 2. mEJP amplitude is reduced in unc-104 mutants and restored in unc-104; wnd double mutants, (related to Figure 3).
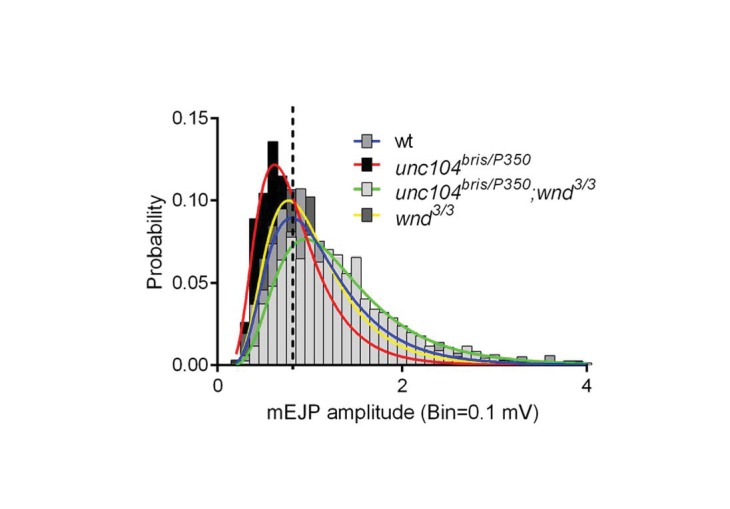
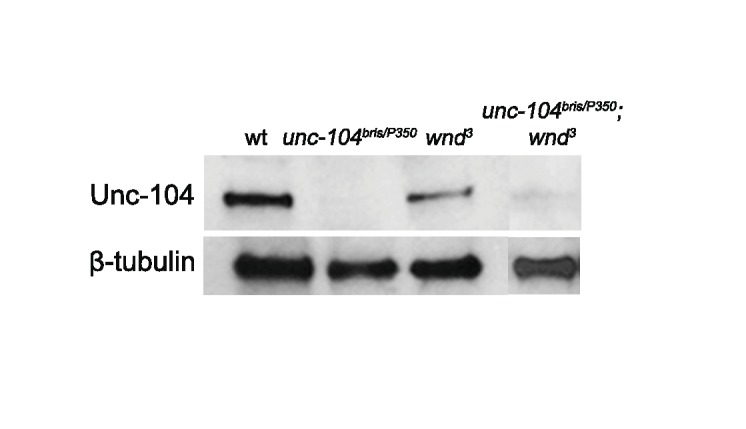
Figure 3—figure supplement 3. The expression of Unc-104 is barely detectable in unc-104-hypomorph mutants, (related to Figure 3).