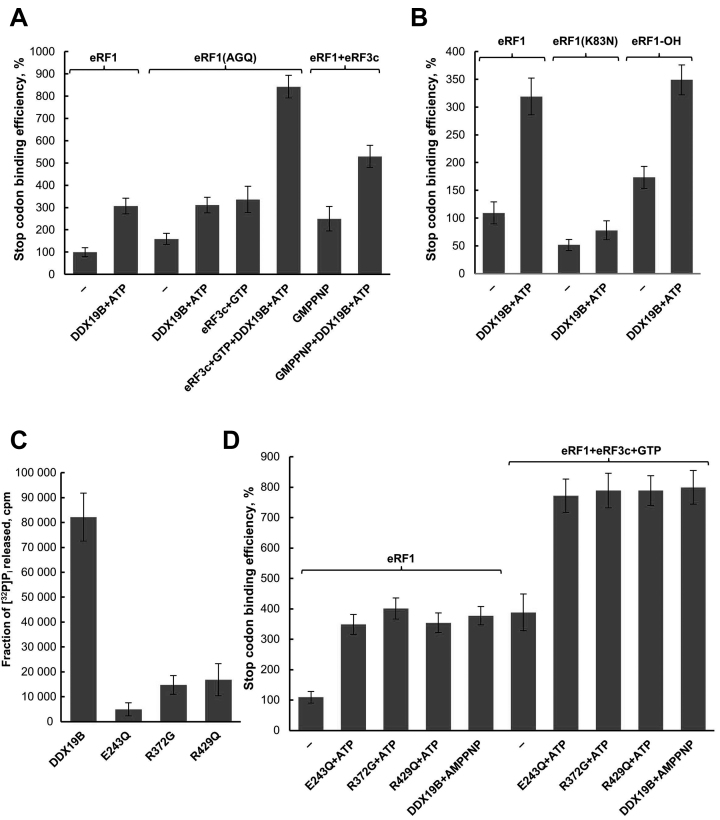
Figure 5.
DDX19B activity in translation termination in the presence of eRFs variants. Relative quantitative analysis of the stop codon binding efficiency of (A) eRF1(AGQ), eRF1(AGQ)+eRF3c+GTP, eRF1+eRF3c+GMPPNP and (B) eRF1(K83N), hydroxylated eRF1 (eRF1-OH) in the presence of DDX19B. Stop codon binding efficiency of eRF1 alone was set as 100% (n = 3). (C) ATPase activity of DDX19B mutants E243Q, R372G and R429Q (n = 3). (D) Relative quantitative analysis of the stop codon binding efficiency of eRF1, eRF1+eRF3c+GTP and DDX19B mutants in the presence of ATP or AMPPNP. Stop codon binding efficiency of eRF1 alone was set as 100% (n = 3). The error bars represent the standard deviation.