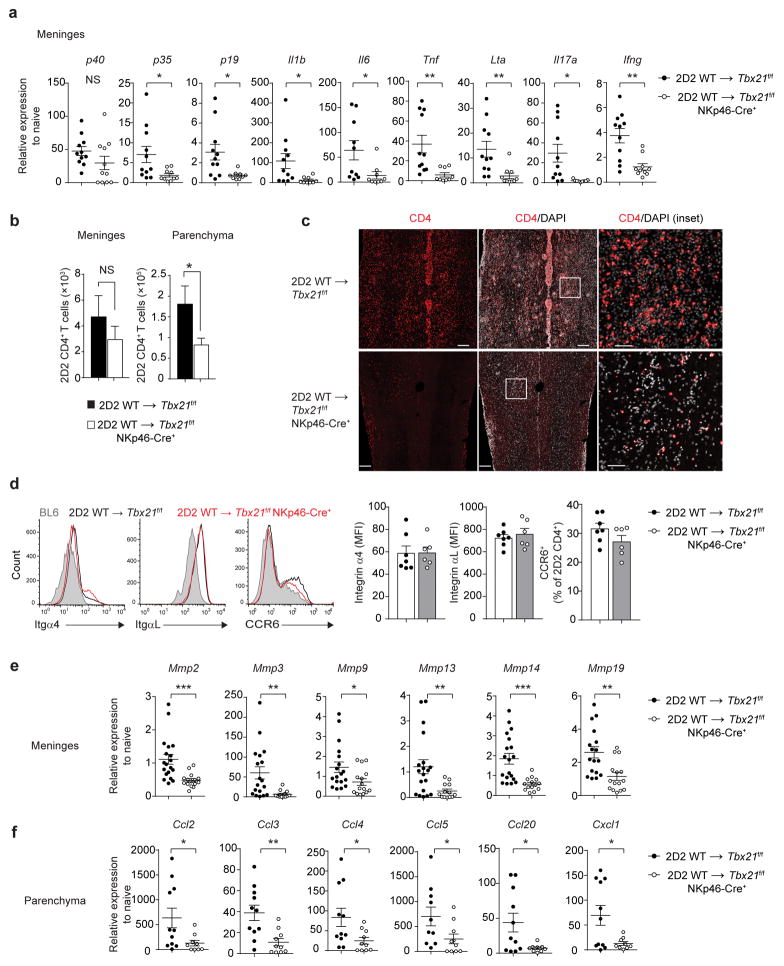
Figure 7.
NKp46+ ILCs regulate infiltration of CD4+ TH17 cells. (a) Quantitative RT-PCR analysis of genes encoding pro-inflammatory cytokines in the spinal cord meninges of Tbx21f/f or Tbx21f/f NKp46-Cre+ mice following the adoptive transfer of 7.5 × 106 2D2 WT TH17 cells. Gene expression is presented relative to the housekeeping gene Hprt, and normalized to transcript abundance from naïve WT (BL6) mice. (b) Absolute number of 2D2 CD4+ T cells (CD4+Vβ11+) in the spinal cord meninges and CNS parenchyma of Tbx21f/f or Tbx21f/f NKp46-Cre+ mice as quantitated by flow cytometry. (c) Immunohistological analysis of longitudinal spinal cord sections from Tbx21f/f or Tbx21f/f NKp46-Cre+ mice following the adoptive transfer of 2D2 WT TH17 cells. Tissue sections were stained with DAPI and anti-CD4. Scale bars, 200 μm for main images or 50 μm for insets. (d) Expression of integrins α4, αL and CCR6 on CNS-infiltrating 2D2 CD4+ T cells (day 18–20 post-adoptive transfer). Bar graphs depict the average mean fluorescence intensity (MFI) of α4 and αL staining and mean percentage of CCR6+ 2D2 CD4+ T cells. (e–f) Quantitative RT-PCR analysis of genes encoding matrix metalloproteinases (MMPs) in the spinal cord meninges and chemokines in the spinal cord parenchyma of Tbx21f/f or Tbx21f/f NKp46-Cre+ mice (quantified as in a). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed Student’s t-test). Data are combined from two independent experiments (a,f; mean ± s.e.m. of n = 11 mice per group, d; n = 6, 7 mice per group), three independent experiments (e; mean ± s.e.m. of n =15, 19 mice per group) or four independent experiments (b; mean ± s.e.m. of 17 mice per group). Histology images are representative of 2 independent experiments (n = 6 mice per group).