Abstract
DNA polymerases catalyze efficient and high fidelity DNA synthesis. While this reaction favors nucleotide incorporation, polymerases also catalyze a reverse reaction, pyrophosphorolysis, removing the DNA primer terminus and generating deoxynucleoside triphosphates. Since pyrophosphorolysis can influence polymerase fidelity and sensitivity to chain-terminating nucleosides, we analyzed pyrophosphorolysis with human DNA polymerase β and found the reaction to be inefficient. The lack of a thio-elemental effect indicated that it was limited by a non-chemical step. Utilizing a pyrophosphate analog, where the bridging oxygen is replaced with an imido-group (PNP), increased the rate of the reverse reaction and displayed a large thio-elemental effect indicating that chemistry was now rate determining. Time-lapse crystallography with PNP captured structures consistent with a chemical equilibrium that favored the reverse reaction. These results highlight the importance of the bridging atom between the β- and γ-phosphates of the incoming nucleotide in reaction chemistry, enzyme conformational changes, and overall reaction equilibrium.
DNA polymerases synthesize DNA during replication and repair of the genome1. Accordingly, they are an attractive target for chemotherapies for uncontrolled cell growth; e.g., cancer and viral infections. There are at least 17 human DNA polymerases that utilize a common nucleotidyl transferase reaction where a deoxynucleoside triphosphate (dNTP) is added to the 3′-end of a growing DNA primer in a template-dependent manner. The reaction requires at least two divalent metal ions that facilitate an inline nucleophilic attack of the primer 3′-oxyanion on Pα of the incoming dNTP resulting in the primer strand extended by one nucleotide (i.e., dNMP) and pyrophosphate (PPi). This reaction is reversible so that PPi and DNA can generate dNTP and a DNA primer strand that is one nucleotide shorter and is termed pyrophosphorolysis2.
Although the forward DNA synthesis reaction is purposely favored, pyrophosphorolysis can be biologically important. Chain-terminating nucleoside drugs are often utilized in an attempt to block DNA synthesis3,4. However, drug resistance to chain-terminating agents can be correlated with an ability of stalled DNA polymerase to remove these nucleotides through pyrophosphorolysis5–7. Additionally, pyrophosphorolysis can remove mis-inserted nucleotides opposite some DNA lesions as a proofreading activity thereby increasing lesion-bypass fidelity8.
DNA polymerase (pol) β is a model DNA polymerase for computational, structural, kinetic, and biological studies9,10. The pyrophosphorolysis activity of pol β is highly dependent on the nature of the DNA substrate11,12. For pyrophosphorolysis, productive substrate binding requires that the primer 3′-terminus be bound in the nucleotide-binding pocket. In contrast, DNA synthesis requires that the primer terminus not occlude this site, but be situated at its boundary. These sites are termed the N- (i.e., post-insertion/pre-translocation) and P-sites, respectively13. Structural studies indicate that the primer terminus is preferentially bound in the P-site with one-nucleotide gapped DNA and in the N-site with nicked DNA14. Adding PPi/Mg2+ to crystals of binary complexes of pol β with nicked DNA generates a stable ternary product (pol/DNAnicked/PPi)11. Due to the unfavorable equilibrium for the reverse reaction, the level of pol/DNAgap/dNTP complex would be beyond the limits of structural detection.
Here we have kinetically characterized pyrophosphorolysis and identify a PPi analog, imidodiphosphate (PNP), that alters the internal equilibrium permitting structural characterization by time-lapse X-ray crystallography. While pyrophosphorolysis was limited by a non-chemical step, replacing the bridging oxygen of PPi with an imido-group resulted in a change in rate limiting step so that the PNP-dependent reverse reaction was limited by chemistry. The results impact our mechanistic understanding of DNA polymerase nucleotidyl transferase chemistry and how key enzyme structural transitions can influence function.
RESULTS
Pyrophosphorolysis is a PPi-dependent reverse reaction
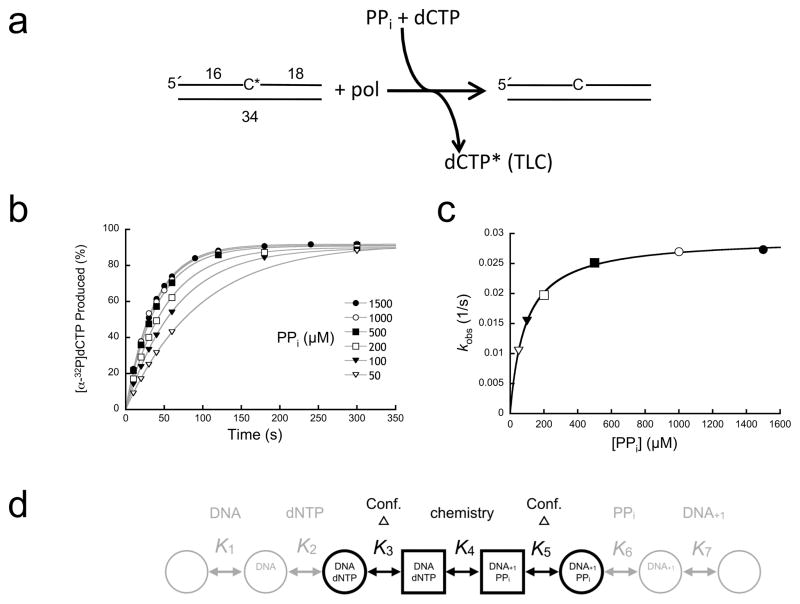
Since nicked DNA positions the primer-terminus at the N-site, a nicked substrate with a 32P-labeled 3′-primer terminus was routinely used for kinetic measurements (Fig. 1a). With this DNA substrate, pyrophosphorolysis generates [α-32P]dNTP and single-nucleotide gapped DNA. Pyrophosphorolysis can be observed by the loss of radioactively labeled DNA or the formation of radiolabeled dNTP. By following the formation of [32P]dCTP with thin-layer chromatography (TLC), the observed rate of the single-turnover (enzyme>DNA) time courses was dependent on PPi concentration (Fig. 1b). A secondary plot of these observed rate constants provided the apparent PPi binding affinity (~90 μM) and observed rate constant for pyrophosphorolysis (krev ~0.03 s−1, Fig. 1c; Supplementary Results, Supplementary Table 1).
Figure 1. Single-turnover analysis of pyrophosphorolysis.
(a) Diagram illustrating the assay used to follow pyrophosphorolysis. A nicked DNA substrate utilizes PPi to remove the 3′-[32P]dCMP (C*) generating [α-32P]dCTP (dCTP*). A cold dCTP trap was included in the reaction to prevent insertion of the radioactive product and to regenerate nicked DNA with an unlabeled 3′-terminus. Product formation (dCTP*) was monitored by TLC. (b) Data points, time, and ligand concentrations were selected to provide full coverage; i.e., multiple points were collected below and above reaction half-times (≥6 time points) and ligand binding affinities (≥5 concentrations), respectively. Time courses were fit to a single exponential (gray lines). (c) A secondary plot of the PPi concentration dependence of the observed first-order rate constants (kobs). These data were fit to a hyperbola (Eq. 1, gray line) to derive krev and Kd (Supplementary Table 1). (d) Simplified kinetic scheme for a DNA polymerase single-nucleotide insertion reaction. The chemical step (K4) is flanked by enzyme conformational changes (K3 and K5). Ligand binding (K1, K2, K6, and K7) occurs to one form of the enzyme (circles) that undergoes a non-chemical conformational change to an alternate form (squares). These conformational states are often described as open or closed forms of the polymerase.
Since ligand (dNTP or PPi) binding results in a structural transition from an open to closed polymerase conformation necessary for catalysis, large global conformational changes occur before and after chemistry (Fig. 1d). To ascertain whether the slow rate of the reverse reaction is limited by chemistry or a non-chemical step, the rate of removal of a 3′-terminal phosphorothioate was determined. Due to different steric, electronic, and metal-binding characteristics of sulfur relative to oxygen, a substantial decrease in rate upon sulfur substitution would suggest that chemistry is rate-limiting. The substrate was enzymatically synthesized using the Sp-diastereomer of (α-S)dATP. Since there is inversion of configuration, the reaction generates nicked DNA with a 3′-terminal Rp-phosphorothioate internucleotide linkage15. In contrast to the forward reaction16,17, there is no phosphorothioate elemental effect observed for pyrophosphorolysis (Supplementary Fig. 1) indicating that chemistry is not rate limiting. In addition, the rate constant for pyrophosphorolysis with T-A in the nick was similar to that measured with G-C.
To determine whether PPi binding could circumvent a kinetic roadblock, an exchange reaction was measured to follow the movement of radioactive-label in [32P]PPi into dNTP. If PPi binding occurs prior to the conformational change (Supplementary Fig. 2a), then the rate of exchange and pyrophosphorolysis will be the same. The rate of the exchange reaction during catalytic cycling was identical to the rate measured by single turnover analysis indicating that PPi binding occurs prior to the conformational change that precedes chemistry (Supplementary Fig. 2b).
Survey of PPi analog-dependent reverse reactions
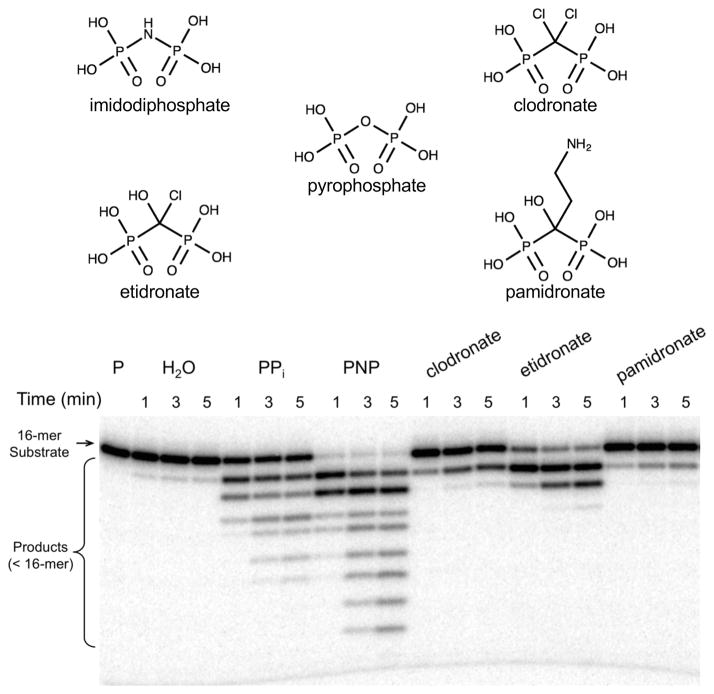
Bisphosphonates (Fig. 2) have a carbon atom in place of the bridging oxygen in PPi and are used to treat osteoporosis and bone metastasis18. We surveyed three bisphosphonates (etidronate, clodronate, and pamidronate) for their ability to serve as substrates for a reverse reaction that would generate a dNTP analog with a modified bridging atom between the β- and γ-phosphates. Additionally, we examined how well imidodiphosphate19, which has been used as part of ATP and GTP analogs (i.e., NMPPNP) to study adenylyl- and guanyl-utilizing enzymes, can serve as a PPi analog for the reverse reaction.
Figure 2. Qualitative assay of pol β reverse reaction with various PPi analogs.
Pol β was pre-incubated with 5′-[32P]-labeled nicked DNA substrate for 5 min at 37 °C and mixed with MgCl2 and PPi or an analog. The final concentrations of MgCl2 and PPi (analog) were 10 and 1 mM, respectively. The full gel is shown in Supplementary Figure 7a. The reverse reaction generates products shorter that the 16-mer-primer. The structures of PPi, imidodiphosphate (PNP), and three bisphosphonates (chlodronate, etidronate, and pamidronate) surveyed are shown above the gel image.
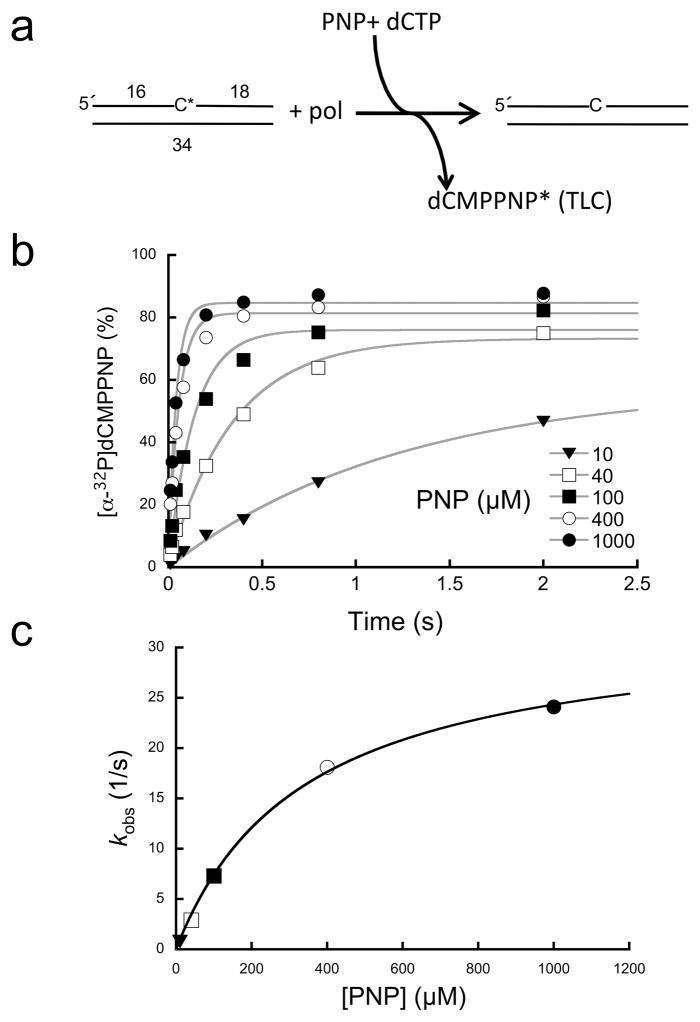
A qualitative assay to survey these analogs indicated that the bisphosphonates were moderate (e.g., etidronate) to poor (chlodronate and pamidronate) substrates, but that PNP exhibited strong activity continually degrading most of the DNA substrate to very short products (Fig. 2). To quantify the impact of substituting nitrogen for the bridging oxygen of PPi, single-turnover experiments were performed to determine the rate and PNP binding affinity of the enzyme (Fig. 3 and Supplementary Table 1). Although the PNP binding affinity was somewhat weaker than PPi (~4-fold), the rate of the reverse reaction was 1000-fold more rapid (krev ~ 30/s).
Figure 3. Single-turnover analysis of PNP-dependent reverse reaction.
(a) Diagram illustrating the assay used to follow the reverse reaction. A nicked DNA substrate utilizes PNP to remove 3′-[32P]dCMP (C*) generating [α-32P]dCMPPNP (dCMPPNP*). A cold dCTP trap was included in the reaction to prevent insertion of the radioactive product and to regenerate nicked DNA with an unlabeled 3′-terminus. Product formation (dCMPPNP*) was monitored by TLC. (b) Data points, time, and ligand concentrations were selected to provide full coverage; i.e., multiple points were collected below and above reaction half-times (≥6 time points) and ligand binding affinities (≥5 concentrations), respectively. Time courses were fit to a single exponential (gray lines). (c) A secondary plot of the PNP concentration dependence of the observed first-order rate constants (kobs). These data were fit to a hyperbola (Eq. 1, gray line) to derive krev and Kd (Supplementary Table 1).
With the TLC solvent system, the dCMPPNP product migrated with a mobility similar to that expected for dCDP as observed previously20. We also verified that the product of the PNP-initiated reverse reaction could be utilized in the forward reaction in a coupled DNA synthesis reaction. This reaction utilized two DNA substrates: unlabeled nicked DNA with a 3′-dCMP at the margin of the nick, and a single-nucleotide gapped DNA with a templating deoxyguanosine in the gap and a 5′-32P-labeled primer. Addition of a low concentration of PNP resulted in gap-filling DNA synthesis on the gapped DNA substrate indicating that PNP can generate dCMPPNP that can subsequently fill the gapped substrate (Supplementary Fig. 3). Additionally, crystallographic characterization of the ternary nicked DNA/PNP complex indicated that PNP supports a strong reverse reaction that generates a dNTP analog where the bridging atom between Pβ and Pγ is a nitrogen atom (see below).
To see whether the faster rate of the observed reverse reaction initiated with PNP is limited by chemistry or a non-chemical step, the rate of removal of a 3′-terminal phosphorothioate was determined. In contrast to the lack of a phosphorothioate elemental effect on the observed reaction initiated with PPi (Supplementary Fig. 1), the PNP-dependent reverse reaction was considerably slower when removing a 3′-terminal Rp-phosphorothioate nucleotide (Supplementary Fig. 4). In this situation, the phosphorothioate effect was ~30.
Since substitution of the bridging oxygen between Pβ and Pγ of dGTP with methylene derivatives has been shown to influence insertion efficiency21–24, the kinetics of 2′-deoxyguanosine-5′-[(β,γ)-imido]triphosphate (dGMPPNP) insertion was quantified. A single turnover analysis indicated that the rate of insertion was strongly diminished, but that binding affinity was increased (Supplementary Fig. 5 and Supplementary Table 1). The decreased forward reaction coupled with the increase in the reverse reaction indicates a decreased chemical equilibrium relative to PPi.
Overall equilibrium constant
The overall equilibrium constant of enzyme-bound gapped DNA and nicked DNA was measured at several PPi concentrations. Under single-turnover conditions, 50 nM gapped DNA was incubated with Mg2+ and a low concentration of dCTP with varying concentrations of PPi. The reactions were quenched after various time intervals and reaction products separated on a sequencing gel. The substrate (gapped DNA) and product (nicked DNA) bands were quantitated, and the overall equilibrium constant calculated (Supplementary Fig. 6); Keq = [nicked DNA][PPi]/[gapped DNA][dCTP]. The overall equilibrium constant determined at different PPi concentrations is Keq = 68,700 ± 7,200.
In the case of the imido-analogs (dGMPPNP and PNP), the overall equilibrium was measured in a similar manner. In this case, the templating base was cytosine since the incoming nucleotide is dGMPPNP. In this situation, the concentration of dGMPPNP required to generate a substantial forward reaction, balancing the strong reverse reaction with PNP, is much higher than when the bridging atom between Pβ and Pγ of the triphosphate is an oxygen atom (Supplementary Fig. 6). The overall equilibrium was considerably lower than with natural substrates (Keq = 1.7 ± 0.1).
DNA substrate specificity for the reverse reaction
Insertion of a chain-terminating nucleotide or an incorrect nucleotide results in a DNA product that disrupts further DNA synthesis. This provides an opportunity to remove the 3′-terminal nucleotide by pyrophosphorolysis. The PPi-dependent removal of a 3′-mismatched nucleotide is very poor probably due to the distorted active site geometry of the terminal mismatch in the polymerase active site25. However, chain-terminating nucleotides are often modified in their sugar moiety that does not perturb Watson-Crick hydrogen bonding or phosphate backbone geometry thereby providing a good substrate for an unblocking reaction (i.e., “removal by reversal”).
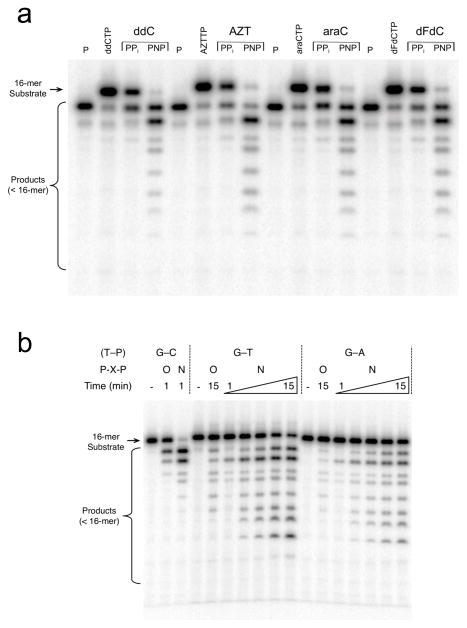
Pyrophosphate- and PNP-dependent removal of chain-terminating nucleotides is shown in Figure 4. The DNA substrates were prepared in situ starting with single-nucleotide gapped DNA substrates where the primer strand is 5′-32P-labeled. An excess (relative to DNA) of chain-terminating nucleoside triphosphate was added to the gapped DNA substrate and incubated with pol β to generate a nicked DNA substrate with a 3′-chain-terminating nucleotide. Pyrophosphate or PNP was added and shortening of the labeled DNA primer strand was monitored. The chain-terminating nucleotides were removed by pyrophosphorolysis, but were more rapidly removed when PNP was substituted for PPi.
Figure 4. Removal of aberrant primer termini by pol β-dependent reverse reaction.
(a) Pol β and one-nucleotide gapped DNA were mixed with MgCl2 and various triphosphates of chain-terminating nucleotides (ddCTP, AZTTP, araCTP, or dFdCTP) as outlined in Online Methods. The gap filling reaction generated a nicked DNA substrate. The reverse reaction was initiated by addition of MgCl2 and PPi or PNP. After 3 min, an aliquot was removed, quenched, and analyzed on a denaturing gel. The 15-mer primer (P), 16-mer terminated nicked DNA substrate (ddCMP, AZTMP, araCMP, or dFdCMP) and reverse reaction products (<16-mer) are indicated. The full gel is shown in Supplementary Figure 7b. (b) Pol β was pre-incubated with 5′-[32P]-labeled nicked DNA substrate with a matched (G-C) or mismatched (G-A or G-T) primer terminal base pair and mixed with mM MgCl2 and PPi or PNP. The 16-mer substrate and reverse reaction products (<16-mer) are indicated. The full gel is shown in Supplementary Figure 7c.
In contrast to Watson-Crick base-paired primer termini, mismatches at the primer terminus are not good substrates for pyrophosphorolysis11. However, substituting PNP for PPi resulted in substantial removal of the mismatch (Fig. 4). Additionally, it appears that a G-T mismatch (template-primer) was removed more rapidly than a G-A terminus.
Time-lapse crystallography of pyrophosphorolysis
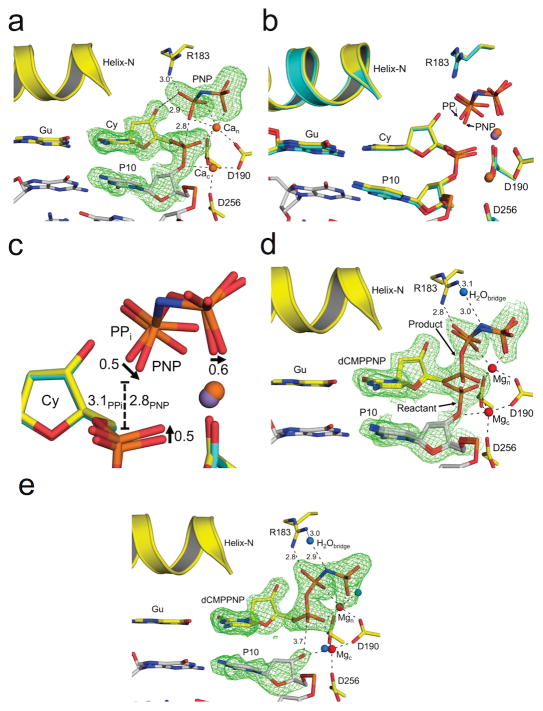
To analyze the robust nature of the reverse reaction in molecular detail, time-lapse crystallography was performed. In this approach, crystals of binary DNA complexes are soaked with substrates or metals to initiate the chemical reaction and the reaction stopped at time intervals by rapid freezing. The structure is then determined to identify the progress of the reaction and capture unique molecular aspects along the reaction path. Although this has been accomplished for the forward DNA synthesis reaction, it was not successful in initiating pyrophosphorolysis; i.e., PPi binding did not generate dNTP. This is probably due the unfavorable chemical equilibrium. Since PNP-initiated a strong reverse reaction, PNP/Ca2+ was added to crystals of binary pol β/nicked DNA complexes. Because Ca2+ is catalytically inert, PNP binding resulted in a closed pre-catalytic ternary complex (pol/DNAnicked/PNP) with two Ca2+ ions positioned in the metal binding sites necessary for the forward DNA synthesis reaction (Supplementary Table 2 and Fig. 5a). The ternary PNP/Ca2+ complex was compared to the ternary PPi product complex generated with Mg2+ (PDB 4KLO; Fig. 5b)11. While the structures are globally similar (RMSD = 0.29 Å over 326 Cα atoms), there are several notable and subtle differences. The PNP structure included two Ca2+ ions in the catalytic and nucleotide metal binding sites, whereas the PPi structure has a single Mg2+ ion bound to the nucleotide metal site and a Na+ bound in the catalytic metal site. Additionally, the precise position of the non-bridging oxygens of PNP are shifted ~0.5 Å relative to that observed with PPi (Fig. 5c). This modest re-positioning moves the attacking oxygen on PNP 0.3 Å nearer the phosphate of the leaving group as compared to PPi (2.8 and 3.1 Å, respectively).
Figure 5. Observing the reverse reaction by time-lapse crystallography.
The pol β active site is shown with key residues indicated; all Fo –Fc omit maps are contoured at 3σ (green). Metal coordination and key distances (Å) are indicated with dashed lines. The carbons of the terminal base pair of the nicked DNA are yellow. The carbons of the upstream DNA are gray. The primer nucleotide upstream of the primer terminus (P10), as well as PNP are indicated. The bridging nitrogen of PNP is colored blue. (a) The active site for the ground state nicked DNA substrate complex with PNP and Ca2+ (orange) is shown. (b) An overlay of the substrate nicked DNA/PNP/Ca2+ complex (yellow carbons) and the nicked DNA/PPi/Mn2+ product complex (PDB code 4KLH; light blue carbons) is shown. The manganese atom from the PPi complex is purple. (c) A close-up of the PPi and PNP phosphate groups from b. The arrows indicate the phosphate oxygen shift for PNP relative to PPi. The distance between the phosphate and the attacking oxygen for PNP and PPi is indicated with a dashed line. (d) The reactant complex for the reverse reaction is shown following a short MgCl2 soak. The Mg2+ and water ions are shown as red and blue spheres, respectively. The distances between the bridging water, Arg183, and the nitrogen of PNP are indicated. The catalytic and nucleotide binding metals are labeled as Mgc and Mgn, respectively. (e) The final one-nucleotide gapped DNA/dCMPPNP ternary complex is shown following the reverse reaction.
Soaking the crystals in a solution containing Mg2+ resulted in Mg2+exchange for the Ca2+ ions. After a short time, the crystals were flash frozen and diffracted to 2.0 Å. Occupancy refinement indicated that approximately 40% of the complexes had undergone a reverse reaction. The catalytic and metal binding sites contained Mg2+ ions as deduced by coordination distances and geometry. Additionally, a unique water molecule serves as a ‘bridging’ molecule between Arg183 and the nitrogen between Pβ and Pγ of dCMPPNP (Fig. 5d). Other Mg2+ soaks resulted in complete turnover of the crystallographic complexes with product dCMPPNP bound in the closed polymerase complex. Notably, Mg2+ still occupies the catalytic metal site without apparent DNA synthesis activity. In this case, the distance between O3′ (primer terminus) and Pα (dCMPPNP) is 3.7 Å compared to 3.4 Å observed with deoxyuridine-5′-[(β,γ)-imido]triphosphate (PDB 2FMS, Fig. 5e).
DISCUSSION
Pyrophosphorolysis has been suggested to play a role in DNA polymerase fidelity8,26,27 and HIV-1 reverse transcriptase5, as well as mitochondrial DNA polymerase γ, sensitivity to chain-terminating nucleoside drugs7. DNA polymerases that stall after insertion of a chain-terminating or aberrant nucleotide can utilize pyrophosphorolysis to remove this impediment, whereas DNA polymerases with a proofreading 3′–5′ exonuclease could employ the hydrolytic excision activity to remove the terminal nucleotide. In this latter case, a nucleoside monophosphate instead of the triphosphate is produced. A better understanding of the reverse reaction is essential for defining the overall reaction that will impact or modulate these proposed activities and is a pre-requisite for rational drug design. In this respect, PPi analogs can inhibit the forward or reverse reaction whereas others that enhance the reverse reaction can decrease the overall forward reaction28,29.
The oversimplified general scheme for DNA polymerase single nucleotide insertion (Fig. 1d) serves as a useful outline to discuss and interpret kinetic and structural observations. It does not include several key steps that can have substantial impact on activity such as catalytic metal binding and additional conformational adjustments that would impact the distribution of the enzyme-ligand complexes30. The identities of the pre- and post-chemistry conformational change steps are also not known. However, intensive structural characterization of a wide variety of DNA polymerases in different liganded states indicates that there are protein and substrate conformational adjustments upon ligand binding. These range from large enzyme subdomains motions (e.g., T7 DNA polymerase) to subtle loop and side chain adjustments (e.g., pol μ). Pol β–DNA binary complexes (nicked or gapped DNA) transition to closed complexes when they bind PPi or dNTP. This involves repositioning of the carboxyl-terminal N-subdomain (‘fingers’ of right-handed DNA polymerases) to make intimate contacts with substrates and products. Thus, the opening and closing of the N-subdomain will be used in the context of the conformational changes (Fig. 1d).
Substrate and protein conformational adjustments play an important role facilitating a commitment to high fidelity DNA synthesis by sequestering the correct nucleoside triphosphate (large K3, Fig. 1d) and aligning catalytic atoms31. In addition, rapid decomposition of the ternary product complex through a two-step reaction where a post-chemistry conformational change (large K5) facilitating rapid PPi release also commits the reaction forward. While a two-step dNTP binding mechanism is well established, the impacts of post-chemistry conformational changes and pyrophosphorolysis have received less attention. To analyze kinetic steps that occur after nucleotide insertion, the reverse reaction was characterized.
DNA polymerases have evolved to replicate DNA while deterring the reverse nucleic acid-degrading pyrophosphorolysis reaction. This is partly due to utilizing a highly charged active site that ‘tunes’ natural substrates for DNA synthesis. Experimental estimates for the equilibrium constant with A- and B-family proofreading DNA polymerases (exo– mutants) are ~5000 (refs. 32–35). For pol β (X-family), which lacks a proofreading activity, the equilibrium constant determined from the equilibrium concentration of enzyme bound substrates and products is >10-fold higher. This greater commitment to the forward reaction could be partly due to rapid catalytic metal dissociation after nucleotide insertion observed for pol β that would deter the reverse reaction. Quantum mechanics/molecular mechanics calculations indicate that this metal is required for pyrophosphorolysis36. Additionally, post-catalytic active site water penetration leads to the loss of nucleotide metal coordination with PPi thereby initiating product dissociation11 that would also deter pyrophosphorolysis.
DNA polymerase β pyrophosphorolysis is slow (krev ~0.03 s−1), as measured by single-turnover analysis (enzyme>DNA, no catalytic cycling) as well as an exchange reaction that measures the movement of radiolabel from PPi to dNTP during alternating nucleotide insertion and removal (Supplementary Fig. 2). The lack of a thio-elemental affect for pyrophosphorolysis is consistent with a rate-limiting non-chemical conformational change preceding pyrophosphorolysis (Supplementary Fig. 1a). With an unfavorable equilibrium constant after nucleotide insertion chemistry (large K5), the observed rate for pyrophosphorolysis would underestimate the intrinsic rate (k−4, Fig. 1d) since the productive ternary product complex would only be a fraction of the total enzyme product (DNA+1/PPi) complexes.
Employing nucleoside triphosphates that have modified leaving groups (i.e., bridging β,γ-methylene-derivatives), nucleotide insertion was shown to be strongly dependent on leaving group acidity (lower acidity resulted in decreased insertion) suggesting that PPi bond breaking is at least partially rate limiting21–24. The acidity of β,γ-imido-modified nucleoside triphosphates are lower than their natural counterparts37. In agreement with methylene substitutions, the insertion of dGMPPNP is diminished by 2 orders of magnitude while the observed reverse reaction with PNP is increased by 3 orders of magnitude (Supplementary Table 1), suggesting that the overall equilibrium is altered ~105-fold.
Substitution of sulfur for a non-bridging oxygen atom on Pα (dNTP) provides valuable mechanistic details for the polymerase-catalyzed reactions15,38. Sulfur substitution for a non-bridging oxygen on Pα should make this phosphate less susceptible to nucleophilic attack (sulfur is less electronegative than oxygen), thereby reducing the observed rate if chemistry is the sole rate-limiting step. While there was not a thio-elemental affect with the PPi-initiated reverse reaction, a substantial affect (~30; Supplementary Fig. 4) was observed with the PNP-initiated reaction. The more rapid rate must reflect a substantial increase in the rate of the conformational change that precedes the reverse reaction (k−5) so that it is no longer rate-limiting. The water-mediated hydrogen bonding observed between the imido-moiety and Arg183 may facilitate this step (Fig. 5d,e). A similar hydrogen bonding pattern has been reported with a pol β ternary complex with gapped DNA and thymidine-5′-[(α,β)-methyl:(β,δ)-imido]triphosphate (TMPCPNP)39.
Time-lapse crystallographic characterization of the forward reaction for pol β11 and pol η40 identified an adjunct divalent metal cation coordinating reaction products (i.e., inserted dNMP and PPi). It was proposed that this metal lowers the activation barrier for the insertion reaction (i.e., increases k4)41,42. In contrast, computational studies with pol β are consistent with a role for this metal in deterring the pyrophosphorolysis reaction (i.e., decreases k−4)36. Consistent with the latter interpretation, a closed pol β ternary product complex can be formed with nicked DNA and PPi with an adjunct metal that does not undergo pyrophosphorolysis (i.e., no dNTP formation)11. In addition, we have been unable to solve the structure of a closed binary nicked DNA complex, consistent with rapid PPi release occurring after subdomain opening. Notably, the ability to structurally observe the reverse reaction in the closed complex with PNP, but not PPi, indicates that the internal chemical equilibrium has been dramatically decreased when PPi is substituted with PNP. Importantly, the adjunct product metal that could interfere with the reverse reaction is not observed. The strong thio-elemental effect measured with the PNP-dependent reaction indicates that chemistry is now rate limiting. Thus, at least two steps (Fig. 1d, steps 4 and 5) have been altered to dramatically decrease the equilibrium constant. Upon binding PNP, pol β must close rapidly, forming the activated ternary complex (k−5>k−4). The ability to structurally capture the product complex (i.e., with dCMPPNP and one-nucleotide gapped DNA) of the PNP-dependent reverse reaction with magnesium in the catalytic and nucleotide binding sites, suggests that the chemical equilibrium constant (K4) is substantially less than 1. If the measured single-turnover rates are taken as the intrinsic rate constants for this step, then K4 = 0.003. Since Keq is 1000-fold greater than this, surrounding equilibria must pull the DNA synthesis reaction forward. The distance between the newly formed primer terminus (O3′) and Pα of dCMPPNP (3.7 Å; Fig. 5e) is substantially greater than that observed in a pre-catalytic complex for the forward reaction trapped with a non-hydrolyzable nucleotide analog (3.4 Å)43. This may in part account for the diminished rate of nucleotide insertion.
Pyrophosphate-dependent primer terminus removal is considerably better with a matched than with a mismatched terminus11. Although PNP improves mismatch removal, it is not as good as with a matched terminus (Fig. 4b) indicting that a well-positioned primer terminus is required for optimal activity. The observation that pol λ can remove a misinserted dAMP opposite 8-oxo-deoxyguanosine (8-oxodG) through pyrophosphorolysis is consistent with this idea8. In this context, the mismatch mimics an A-T base pair where 8-oxo-dG is in a syn-conformation Hoogsteen base pairing with adenine.
With natural substrates, resistance to chain-terminating nucleotides is minimized through a post-insertion conformational change that pulls the reaction forward (Fig. 1d, large K5). The reversal of this conformational change (k−5) limits pyrophosphorolysis. In instances where pyrophosphorolysis is elevated, drug resistance can be attributed to an altered post-nucleotide insertion non-chemical step. The molecular identity of this step is unknown, but has often been attributed to subdomain repositioning (opening and closing) known to occur with HIV-1 reverse transcriptase, pol γ, and pol β. Imido-substitution for the β,γ-bridging oxygen in the incoming nucleoside triphosphate and PPi strongly diminished the favorable equilibrium for DNA synthesis by decreasing the forward rate and hastening the reverse reaction (Supplementary Table 1). This occurs by altering conformational and chemical equilibria. Accordingly, the product of the reverse reaction (dGMPPNP) is a good inhibitor of the forward reaction (i.e., binds tightly and is inserted slowly). While PNP-dependent removal of chain-terminating nucleotides is substantially better than with PPi (Fig. 4a), the net result would be very low DNA polymerase activity (i.e., inhibition). Importantly, the equilibrium for the overall reaction is sensitive to the nature of the DNA synthesis leaving group indicating that the chemistry of the terminal phosphates of an incoming nucleotide influences both chemical and conformational equilibria.
ONLINE METHODS
Materials
Human pol β was expressed and purified44. Chlodronate, etidronate, imidodiphosphate, pamidronate, and pyrophosphate were from Sigma-Aldrich. The β,γ-imido modified nucleoside triphosphate analog, 2′-deoxyguanosine-5′-[(β,γ)-imido]triphosphate (dGMPPNP), was from Jena Bioscience. Chain terminating nucleoside triphosphates; ddCTP was from GE Healthcare, 3′-azido-2′,3′-dideoxythymidine triphosphate (AZTTP) and arabinofuranosylcytosine triphosphate (araCTP) were from Trilink Biotechnologies, and gemcitabine (dFdCTP) was obtained from Jena Bioscience. [α-35S]dATP, [α-32P]dCTP, and [32P]PPi were from Perkin Elmer. Polyethyleneimine (PEI) cellulose thin layer chromatography (TLC) plates containing a fluorescent indicator were purchased from EMD Millipore.
Reaction buffer
All kinetic measurements were performed in a buffer containing 50 mM MES, 25 mM Tris, 25 mM ethanolamine (pH 7.5 adjusted at 37 °C), 100 mM KCl, 10 mM MgCl2 supplemented with 10% glycerol, 100 μg/ml bovine serum albumin, 1 mM dithiothreitol, and 0.1 mM EDTA.
Product separation
Changes in the length of a 5′-labeled primer strand were visualized and resolved on 16% denaturing polyacrylamide gels. The gel was scanned using a phosphorimager in fluorescence mode to visualize 6-carboxyfluorescein (6-FAM)-labeled oligonucleotides. Radiolabeled oligonucleotides were detected after exposing a dried gel to a phosphor screen. Reverse reaction products were also separated on PEI cellulose TLC plates. Unless otherwise noted, the plates were developed in 0.2 or 0.3 M NaPi, pH 7.0. Thin layer chromatography of [35S]-labeled reverse reaction products was performed in buffer containing 10 mM β-mercaptoethanol.
DNA preparation
Single nucleotide gapped DNA substrates containing a 5′-6-FAM label were prepared as detailed previously45. Nicked DNA substrates utilized to qualitatively monitor the reverse reaction were prepared as follows. Briefly, a 16-mer oligonucleotide primer was radiolabeled at the 5′-end with [γ-32P]ATP and Optikinase. Unincorporated [γ-32P]ATP was removed using a BioSpin 6 column. The 5′-labeled primer (1 equivalent) was mixed with 1.2 equivalents of 34-mer template and 18-mer downstream oligonucleotide containing a 5′-PO4 group. Annealing was performed in a PCR thermocycler. Oligonucleotides were denatured at 95 °C for 5 min followed by slow cooling (1 °C/min) to 10 °C. The following sequences were used to construct the nicked DNA substrates with a matched or mismatched primer terminus; primer, 5′-CTG CAG CTG ATG CGC Y-3′, where Y denotes A, C or T; downstream oligonucleotide, 5′-GTA CGG ATC CCC CGG GTA C-3′; template strand, 5′-GTA CCC GGG GAT CCG TAC XGC GCA TCA GCT GCA G-3′, where X denotes G.
PNP-induced gap-filling reaction
Pol β (5 μM) was pre-incubated with 2.5 μM nicked DNA and 20 μM PNP in reaction mixture without Mg2+. This was mixed (1:1, v/v) with a solution with 20 mM MgCl2 and incubated at 37 °C in reaction buffer. Following mixing, the final concentrations were 2.5 μM pol β, 1.25 μM nicked DNA, 10 μM PNP and 10 mM MgCl2. After 10 min, an aliquot was mixed (4:1, v/v) with a solution containing 2.5 μM single-nucleotide gapped DNA (G in the gap) with a 5′-6-FAM labeled 15-mer primer and 10 mM MgCl2. Aliquots (10 μl) were removed at various times and quenched in an equal volume of 0.3 M EDTA, pH 8.0. Reaction substrates/products were separated on 16% denaturing polyacrylamide gels and visualized by phosphorimagery.
Preparation of 3′-[32P] or [35S] labeled nicked DNA substrates
DNA polymerase β was used to fill a 1-nucleotide gapped DNA substrate with either [32P]dCTP or [35S]dATP to create a 3′-radiolabeled nicked DNA substrate. The reaction mixture contained 50 mM Tris-Cl, pH 7.4 (37 °C), 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 2.5 μM gapped DNA, 5 μM [32P]dCTP or [35S]dATP. The single-nucleotide DNA substrate was similar to the nicked substrate described above except the primer strand was one nucleotide shorter (3′-nucleotide deleted). Gap filling was initiated by addition of pol β and incubated at 37 °C for 5–10 min. The reaction was quenched by addition of 0.5 M EDTA (0.1 vol). To remove enzyme and unincorporated nucleotides, the mixture was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) followed by two passages through BioSpin 6 columns. Aliquots of the labeling reaction were removed prior to and following the extraction/removal steps to determine final DNA substrate concentration. Pre- and post-aliquots (1 μl) were spotted onto PEI cellulose plates and developed in 0.375 M KH2PO4, pH 4.0. The ratio (post/pre-extraction) was used to correct the initial DNA concentration for loss or dilution of substrate.
Reverse reaction assay
Pol β (1 μM) was pre-incubated with 100 nM nicked DNA containing either a matched or mismatched primer terminus for 5 min at 37 °C in reaction buffer. A solution with 20 mM MgCl2 containing 2 mM PPi or pyrophosphate analog in reaction buffer was used to initiate the reaction. Following mixing, the final concentrations were 500 nM pol β, 50 nM nicked DNA, 10 mM MgCl2, and 1 mM PPi or pyrophosphate analog. Aliquots (5 or 10 μl) were removed at various times and quenched in an equal volume of 0.3 M EDTA, pH 8.0. Reaction substrates/products were separated and visualized as described above.
The removal of a terminated primer terminus by the reverse reaction required enzymatic synthesis of the nicked DNA substrate. A pre-incubated mixture of 4 μM pol γ and 0.4 μM one-nucleotide gapped DNA was mixed 1:1 (v/v) with 20 mM MgCl2 and 0.2 μM various triphosphates of chain-terminating nucleotides (ddCTP, AZTTP, araCTP, or dFdCTP). The gap filling reaction proceeded at 37 °C for 10–20 min to generate a terminated nicked DNA substrate. An aliquot was removed and quenched to verify complete gap filling (16-mer). The reverse reaction was initiated by addition of an equal volume of 10 mM MgCl2 and 250 μM PPi or PNP. An aliquot (time = 3 min) was removed, quenched, and analyzed on a denaturing gel.
The removal of a terminal mismatch by a reverse reaction was followed by incubating 1 μM Pol β with 100 nM 5′-[32P] labeled nicked DNA substrate with a matched (G–C) or mismatched (G-A or G-T) primer terminal base pair for 5 min at 37 °C. This was mixed with a solution of 20 mM MgCl2 and 2 mM PPi or PNP (1:1, v/v) to initiate the reaction. Reactions were quenched at various time intervals with addition of an equal volume of 0.3 M EDTA and substrate and reverse reaction products separated on a denaturing gel.
Kinetic parameters for the reverse reaction were determined under single turnover conditions (E/DNA = 10) at 37 °C. Pol β (1 μM) was pre-incubated with 100 nM 3′-32P-labeled primer in nicked DNA with various concentrations of PPi or PNP in reaction buffer. Time courses were initiated by mixing with an equal volume of a 20 mM MgCl2/50 μM dNTP trap solution in reaction buffer. The dNTP trap prevents re-insertion of radiolabeled product dNTP and corresponds to the identity of the nucleotide triphosphate produced during the reaction. Initiation of the reaction was performed by manual mixing, in the case of pyrophosphorolysis, or rapid mixing using a Kintek RQF-3 with PNP. EDTA (0.1 or 0.2 M) was used as the quenching agent. Substrates/products were resolved by TLC in either 0.2 or 0.3 M NaPi, pH 7.0 buffer.
Pyrophosphate exchange assay
Pol β (2.5 μM) was pre-incubated with 500 nM unlabeled nicked DNA substrate containing a matched primer terminal base pair (G–C, template–primer) in reaction buffer with 20 mM MgCl2 and manually mixed (1:1, v/v) with a pre-warmed solution of reaction buffer, 2 mM [32P]PPi, and 100 μM dCTP. Aliquots were withdrawn at various time points and quenched with 1 vol. of 0.3 M EDTA. Quenched reactions mixtures were applied to PEI cellulose plates and developed in 0.3 M potassium phosphate buffer, pH 8.0. Plates were scanned followed by quantitation using a phosphorimager and ImageQuant software.
Gap filling DNA synthesis kinetic assay
To measure the rate of the first insertion (kpol) and apparent equilibrium nucleotide dissociation constant (Kd), single-turnover kinetic assays (enzyme/DNA = 10) were performed as described previously46. Briefly, a pre-incubated solution of enzyme/DNA was rapidly mixed with various concentrations of MgCl2/dGMPPNP using a Kintek RQF-3 rapid quench-flow. Reactions were quenched with 0.25 M EDTA.
Kinetic analysis
Single-turnover time courses were fit to a single exponential equation to yield the first-order rate constants (kobs) at a given concentration of dGMPPNP, PPi or PNP. Under these conditions, kobs was dependent on the concentration of substrate. A secondary plot of the concentration dependence of kobs was hyperbolic and fitted by non-linear least-squares method to Equation 1 where kmax is the intrinsic rate constant for the step limiting the first nucleotide insertion (forward reaction) or removal (reverse reaction).
| (Eq. 1) |
where S = dGMPPNP, PPi, or PNP. For the insertion of dGMPPNP, the secondary plot was fit to a quadratic equation (Eq. 2) due to its high affinity relative to the enzyme concentration.
| (Eq. 2) |
Data points, time and ligand concentrations, were selected to provide full coverage; i.e., multiple points were collected below and above reaction half-times (≥6 time points) and ligand binding affinities (≥5 concentrations), respectively. Unless noted, kinetic constants represent best-fit parameters and their standard error.
Overall equilibrium constant determination
A mixture of 500 nM pol β with single-nucleotide gapped DNA (pol/DNA = 10; templating G or C) containing various concentrations of PPi (500–2000 μM) or PNP (20, 50, 100 μM) was mixed with an equal volume of 20 mM MgCl2 containing 60–100 nM dCTP or 50 μM dGMPPNP and incubated at 37 °C for various time intervals. Aliquots (10 μl) were withdrawn at various times and quenched with an equal volume of 0.3 M EDTA. The reactions were quenched after 10–80 s and reaction products separated on a sequencing gel. The substrate (gapped DNA) and product (nicked DNA) bands were quantitated, and the overall equilibrium constant calculated; Keq = [nicked DNA][PPi]/[gapped DNA][dCTP]. The mean and standard error for 6 independent determinations are reported in the text.
Structure determination
Binary complex crystals with nicked DNA were grown as previously described43. The time-lapse crystallography was performed as before11 and is briefly summarized here. Binary pol β/DNA complex crystals were first transferred to a cryosolution containing 15% ethylene glycol, 50 mM imidazole, pH 7.5, 20% PEG3350, 90 mM sodium acetate, 2 mM PNP and 50 mM CaCl2 for 1 h. These ground state (GS) ternary complex crystals were then transferred to a cryosolution containing 200 mM MgCl2 for varying times. All reactions were stopped by freezing the crystals at 100K prior to data collection at the home source, 1.54 Å, or the Advanced Photon Source, 1.0 Å (Argonne National Laboratory). In house data collection was done on a SATURN92 CCD detector system mounted on a MiraMax-007HF rotating anode generator. This allows for anomalous data detection after phasing by molecular replacement. Remote data collection was done at Southeast Regional Collaborative Access Team (SER-CAT) BM-22 beamline at the Advanced Photon Source (Argonne National Laboratory) with the MAR225 area detector. Data were processed and scaled using the HKL2000 software package47. Initial models were determined using molecular replacement with the open binary (PDB ID 3ISB) or closed ternary (PDB ID 2FMS) structures of pol β and all Rfree flags were taken from the starting model. Refinement was carried out using PHENIX and model building using Coot48,49. The metal-ligand coordination restraints were generated by ReadySet (PHENIX) and not utilized until the final rounds of refinement. Partial catalysis models were generated with both the reactant and product species and occupancy refinement was performed. The structural figures were prepared in Pymol (Schrödinger, LLC) and all density maps were generated after performing simulated annealing. Ramachandran analysis determined 100% of non-glycine residues lie in allowed regions and at least 97% in favored regions.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: ground state complex (5UGN), reactant state complex (5UGO), and product sate complex (5UGP). All data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank the Collaborative Crystallography group at NIEHS for help with data collection and analysis. Use of the advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (project numbers Z01-ES050158 and Z01-ES050161) (S.H.W.) and was in association with the National Institutes of Health grant 1U19CA105010 (S.H.W.) and R00-ES024431 (B.D.F.).
Footnotes
Author contributions
D.D.S., B.D.F., W.A.B., and S.H.W. designed the project. D.D.S. did the kinetic analyses. B.D.F. carried out crystallography. D.D.S., B.D.F., W.A.B., and S.H.W. prepared the manuscript. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Prot Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher MP, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid XXVIII.Pyrophosphate exchange and pyrophosphorolysis reactions of deoxyribonucleic acid polymerase. J Biol Chem. 1969;244:3019–3028. [PubMed] [Google Scholar]
- 3.Parsons JL, Nicolay NH, Sharma RA. Biological and therapeutic relevance of nonreplicative DNA polymerases to cancer. Antioxid Redox Signal. 2013;18:851–873. doi: 10.1089/ars.2011.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna CE, Kashemirov BA, Peterson LW, Goodman MF. Modifications to the dNTP triphosphate moiety: From mechanistic probes for DNA polymerases to antiviral and anti-cancer drug design. Biochim Biophys Acta, Proteins Proteomics. 2010;1804:1223–1230. doi: 10.1016/j.bbapap.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Smith AJ, Meyer PR, Asthana D, Ashman MR, Scott WA. Intracellular substrates for the primer-unblocking reaction by human immunodeficiency virus type 1 reverse transcriptase: Detection and quantitation in extracts from quiescent- and activated-lymphocyte subpopulations. Antimicrob Agents Chemother. 2005;49:1761–1769. doi: 10.1128/AAC.49.5.1761-1769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urban S, Urban S, Fischer KP, Tyrrell DL. Efficient pyrophosphorolysis by a hepatitis B virus polymerase may be a primer-unblocking mechanism. Proc Natl Acad Sci USA. 2001;98:4984–4989. doi: 10.1073/pnas.091324398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanes JW, Johnson KA. A novel mechanism of selectivity against AZT by the human mitochondrial DNA polymerase. Nucleic Acids Res. 2007;35:6973–6983. doi: 10.1093/nar/gkm695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crespan E, Maga G, Hubscher U. A new proofreading mechanism for lesion bypass by DNA polymerase-λ. EMBO Rep. 2012;13:68–74. doi: 10.1038/embor.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Biochemistry. 2014;53:2768–2780. doi: 10.1021/bi500139h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera L, Beard WA, Pedersen LG, Wilson SH. Chapter Four - Applications of Quantum Mechanical/Molecular Mechanical Methods to the Chemical Insertion Step of DNA and RNA Polymerization. In: Christo ZC, editor. Adv Protein Chem Struct Biol. Vol. 97. Academic Press; 2014. pp. 83–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby TW, et al. Metal-induced DNA translocation leads to DNA polymerase conformational activation. Nucleic Acids Res. 2012;40:2974–2983. doi: 10.1093/nar/gkr1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das K, et al. Conformational states of HIV-1 reverse transcriptase for nucleotide incorporation vs pyrophosphorolysis—binding of foscarnet. ACS Chem Biol. 2016;11:2158–2164. doi: 10.1021/acschembio.6b00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawaya MR, Prasad P, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein F. Nucleoside phosphorothioates. Ann Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 16.Vande Berg BJ, Beard WA, Wilson SH. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β: Implication for the identity of the rate-limiting conformational change. J Biol Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Tsai MD. DNA Polymerase β: Pre-steady-state kinetic analyses of dATPαS stereoselectivity and alteration of the stereoselectivity by various metal ions and by site-directed mutagenesis. Biochemistry. 2001;40:9014–9022. doi: 10.1021/bi010646j. [DOI] [PubMed] [Google Scholar]
- 18.Lipton A. Emerging role of bisphosphonates in the clinic: Antitumor activity and prevention of metastasis to bone. Cancer Treat Rev. 2008;34(Supplement 1):S25–S30. doi: 10.1016/j.ctrv.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Rozovskaya T, et al. Pyrophosphate analogues in pyrophosphorolysis reaction catalyzed by DNA polymerases. FEBS Lett. 1989;247:289–292. doi: 10.1016/0014-5793(89)81354-7. [DOI] [PubMed] [Google Scholar]
- 20.Penningroth SM, Olehnik K, Cheung A. ATP formation from adenyl-5′-yl imidodiphosphate, a nonhydrolyzable ATP analog. J Biol Chem. 1980;255:9545–9548. [PubMed] [Google Scholar]
- 21.Oertell K, et al. Transition state in DNA polymerase β catalysis: Rate-limiting chemistry altered by base-pair configuration. Biochemistry. 2014;53:1842–1848. doi: 10.1021/bi500101z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oertell K, et al. Effect of β,γ-CHF- and β,γ-CHCl-dGTP halogen atom stereochemistry on the transition state of DNA polymerase β. Biochemistry. 2012;51:8491–8501. doi: 10.1021/bi3010335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sucato CA, et al. Modifying the β,γ leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase β. Biochemistry. 2007;46:461–471. doi: 10.1021/bi061517b. [DOI] [PubMed] [Google Scholar]
- 24.Sucato CA, et al. DNA polymerase β fidelity: Halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry. 2008;47:870–879. doi: 10.1021/bi7014162. [DOI] [PubMed] [Google Scholar]
- 25.Batra Vinod K, Beard William A, Pedersen Lars C, Wilson Samuel H. Structures of DNA polymerase mispaired DNA termini transitioning to pre-catalytic complexes support an induced-fit fidelity mechanism. Structure. 2016;24:1863–1875. doi: 10.1016/j.str.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li A, Gong S, Johnson KA. Rate-limiting pyrophosphate release by HIV reverse transcriptase improves fidelity. J Biol Chem. 2016;291:26554–26565. doi: 10.1074/jbc.M116.753152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruchaga C, Ansó E, Rouzaut A, Martínez-Irujo JJ. Selective excision of chain-terminating nucleotides by HIV-1 reverse transcriptase with phosphonoformate as substrate. J Biol Chem. 2006;281:27744–27752. doi: 10.1074/jbc.M603360200. [DOI] [PubMed] [Google Scholar]
- 29.Yanvarev DV, et al. Methylene bisphosphonates as the inhibitors of HIV RT phosphorolytic activity. Biochimie. 2016;127:153–162. doi: 10.1016/j.biochi.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Balbo PB, Wang ECW, Tsai MD. Kinetic mechanism of active site assembly and chemical catalysis of DNA polymerase β. Biochemistry. 2011;50:9865–9875. doi: 10.1021/bi200954r. [DOI] [PubMed] [Google Scholar]
- 31.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow Fragment): Identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 33.Oertell K, et al. Kinetic selection vs. free energy of DNA base pairing in control of polymerase fidelity. Proc Natl Acad Sci USA. 2016;113:E2277–E2285. doi: 10.1073/pnas.1600279113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 35.Olson AC, Patro JN, Urban M, Kuchta RD. The energetic difference between synthesis of correct and incorrect base pairs accounts for highly accurate DNA replication. J Am Chem Soc. 2013;135:1205–1208. doi: 10.1021/ja309866m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera L, et al. Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse. Proc Natl Acad Sci USA. 2015;112:E5228–E5236. doi: 10.1073/pnas.1511207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yount RG. Adenylylimidodiphosphate and guanylylimidodiphosphate. Methods Enzymol. 1974;38:420–427. doi: 10.1016/0076-6879(74)38059-7. [DOI] [PubMed] [Google Scholar]
- 38.Johnson KA. Conformational coupling in DNA polymerase fidelity. Ann Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 39.Kadina AP, et al. Two scaffolds from two flips: (α,β)/(β,γ) CH2/NH “Met-Im” analogues of dTTP. Org Lett. 2015;17:2586–2589. doi: 10.1021/acs.orglett.5b00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura T, Zhao Y, Yamagata Y, Hua Y-j, Yang W. Watching DNA polymerase η make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, Yang W. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016;352:1334–1337. doi: 10.1126/science.aad9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vyas R, Reed AJ, Tokarsky EJ, Suo Z. Viewing human DNA polymerase β faithfully and unfaithfully bypass an oxidative lesion by time-dependent crystallography. J Am Chem Soc. 2015;137:5225–5230. doi: 10.1021/jacs.5b02109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batra VK, et al. Magnesium induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 45.Freudenthal BD, et al. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature. 2015;517:635–639. doi: 10.1038/nature13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beard WA, Shock DD, Batra VK, Prasad R, Wilson SH. Substrate-induced DNA polymerase β activation. J Biol Chem. 2014;289:31411–31422. doi: 10.1074/jbc.M114.607432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processsing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst Sect D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst Sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: ground state complex (5UGN), reactant state complex (5UGO), and product sate complex (5UGP). All data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.