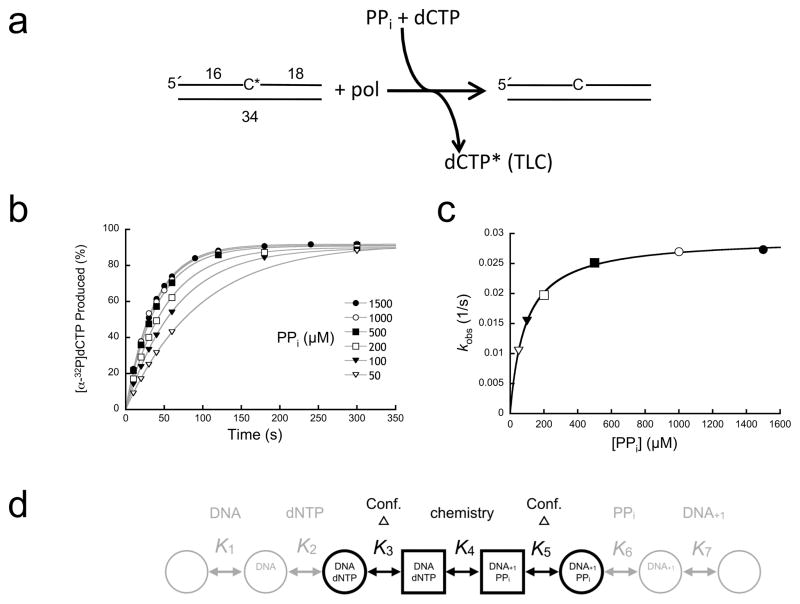
Figure 1. Single-turnover analysis of pyrophosphorolysis.
(a) Diagram illustrating the assay used to follow pyrophosphorolysis. A nicked DNA substrate utilizes PPi to remove the 3′-[32P]dCMP (C*) generating [α-32P]dCTP (dCTP*). A cold dCTP trap was included in the reaction to prevent insertion of the radioactive product and to regenerate nicked DNA with an unlabeled 3′-terminus. Product formation (dCTP*) was monitored by TLC. (b) Data points, time, and ligand concentrations were selected to provide full coverage; i.e., multiple points were collected below and above reaction half-times (≥6 time points) and ligand binding affinities (≥5 concentrations), respectively. Time courses were fit to a single exponential (gray lines). (c) A secondary plot of the PPi concentration dependence of the observed first-order rate constants (kobs). These data were fit to a hyperbola (Eq. 1, gray line) to derive krev and Kd (Supplementary Table 1). (d) Simplified kinetic scheme for a DNA polymerase single-nucleotide insertion reaction. The chemical step (K4) is flanked by enzyme conformational changes (K3 and K5). Ligand binding (K1, K2, K6, and K7) occurs to one form of the enzyme (circles) that undergoes a non-chemical conformational change to an alternate form (squares). These conformational states are often described as open or closed forms of the polymerase.