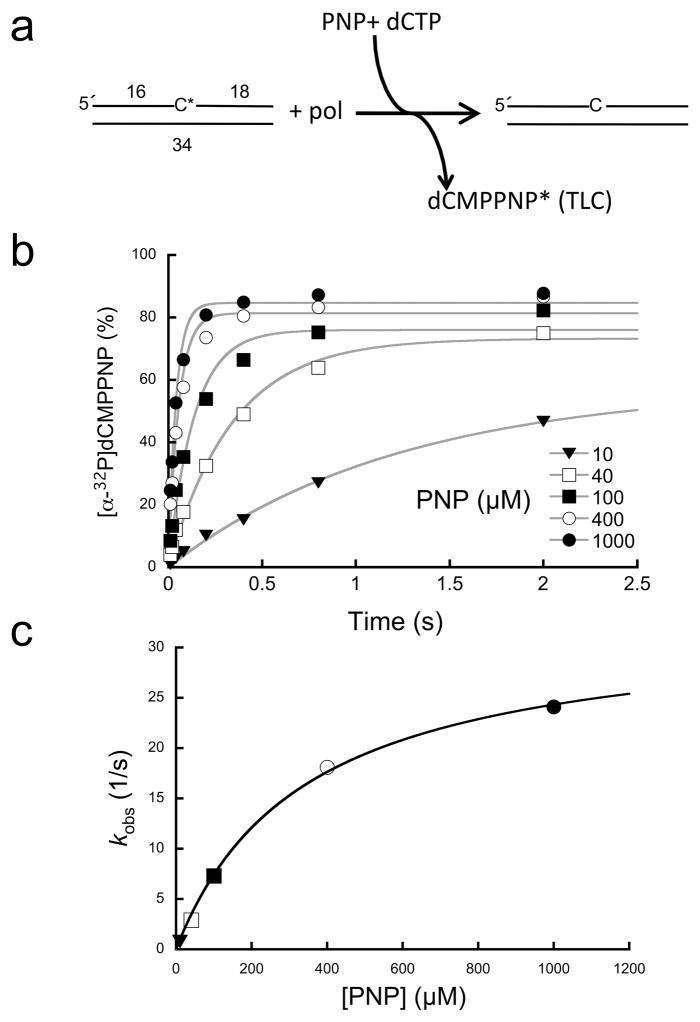
Figure 3. Single-turnover analysis of PNP-dependent reverse reaction.
(a) Diagram illustrating the assay used to follow the reverse reaction. A nicked DNA substrate utilizes PNP to remove 3′-[32P]dCMP (C*) generating [α-32P]dCMPPNP (dCMPPNP*). A cold dCTP trap was included in the reaction to prevent insertion of the radioactive product and to regenerate nicked DNA with an unlabeled 3′-terminus. Product formation (dCMPPNP*) was monitored by TLC. (b) Data points, time, and ligand concentrations were selected to provide full coverage; i.e., multiple points were collected below and above reaction half-times (≥6 time points) and ligand binding affinities (≥5 concentrations), respectively. Time courses were fit to a single exponential (gray lines). (c) A secondary plot of the PNP concentration dependence of the observed first-order rate constants (kobs). These data were fit to a hyperbola (Eq. 1, gray line) to derive krev and Kd (Supplementary Table 1).