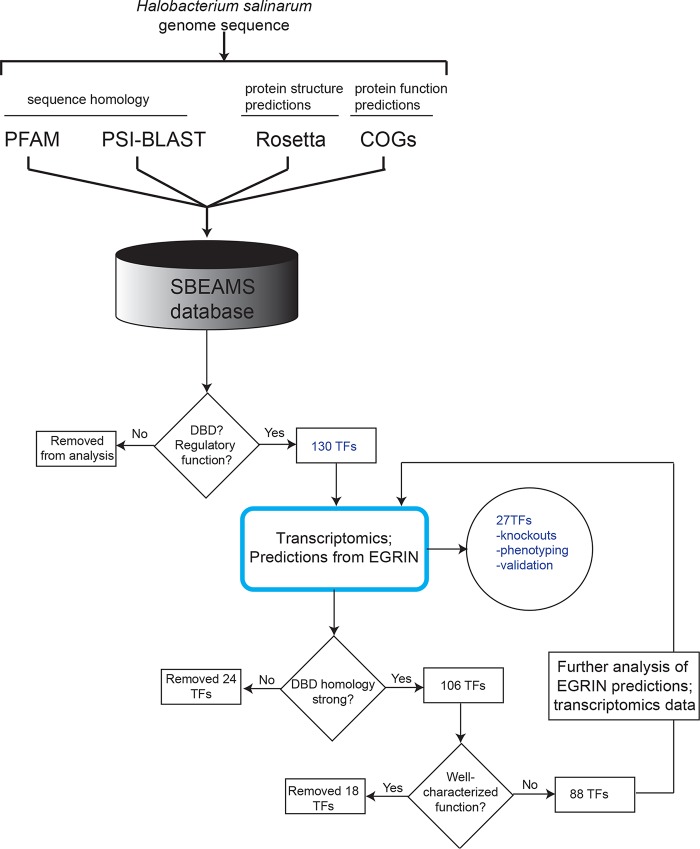
FIG 1 .
TF candidate selection pipeline. Genes encoding proteins with a putative DNA binding domain were annotated using sequence databases PFAM (37) and PSI-BLAST (38), structural predictions (26), and protein functions from COG (39). These annotations were stored in the Systems Biology Experiment and Analysis Management System (SBEAMS) database (36), resulting in 130 putative TFs. Transcriptome analysis across 1,495 experimental conditions (43) and GRN network inference models (7, 15) were then used to generate predictions regarding TF functions. Details of GRN predictions and criteria for selection of the final collection of 27 TFs are given in Table S1 in the supplemental material. DBD, DNA binding domain.