Figure 3. SERRATE regulates hair cell fate and hair length in a partially microRNA-independent manner.
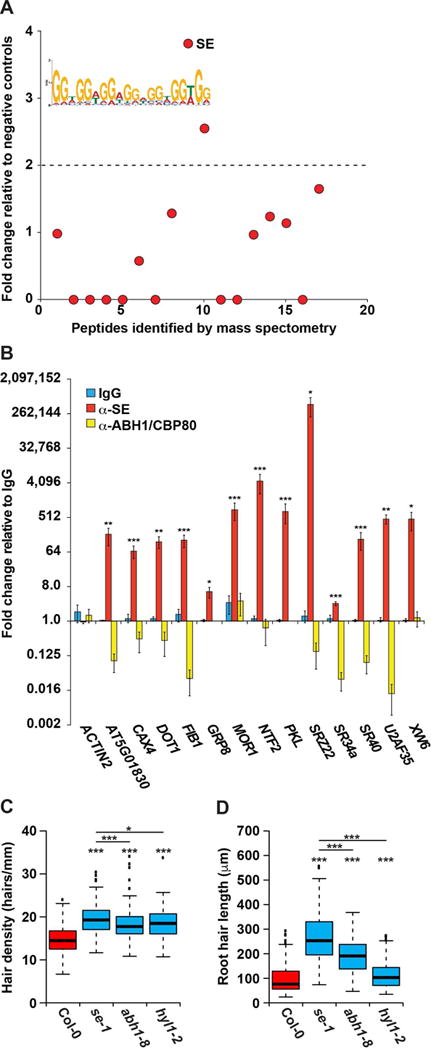
(A) RNA affinity chromatography followed by LC-MS was performed on whole root cell lysate using the MEME identified GGN repeat motif as bait. The number of peptide spectrum matches (PSMs) for each identified peptide was graphed as fold change over the average PSMs of scrambled RNA bait and no RNA controls. Peptides above the dotted line have a more than 2-fold change and correspond to candidate RBPs. SE is denoted as being highly bound by our analysis. (B) RIP-qPCR was performed on whole root lysate using rabbit α-IgG (blue bars), α-SE (red bars), or α-ABH1/CBP80 (yellow bars) antibodies, graphed as fold change relative to the IgG negative control pull down, n = 4. Error bars indicate SEM. *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Welch’s t-test. (C–D) Root hair cell density (hairs/mm) (C) and root hair length (μm) (D) of Col-0, se-1, abh1-8, and hyl1-2 mutant plants. For analysis of root hair length n=400, and for root hair density n > 135. *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, while N.S. denotes p value > 0.05, Wilcoxon test. See also Figures S1, S2, and S4–S6.
