Figure 5. GRP8 regulates root hair cell fate in a GRP7-independent manner.
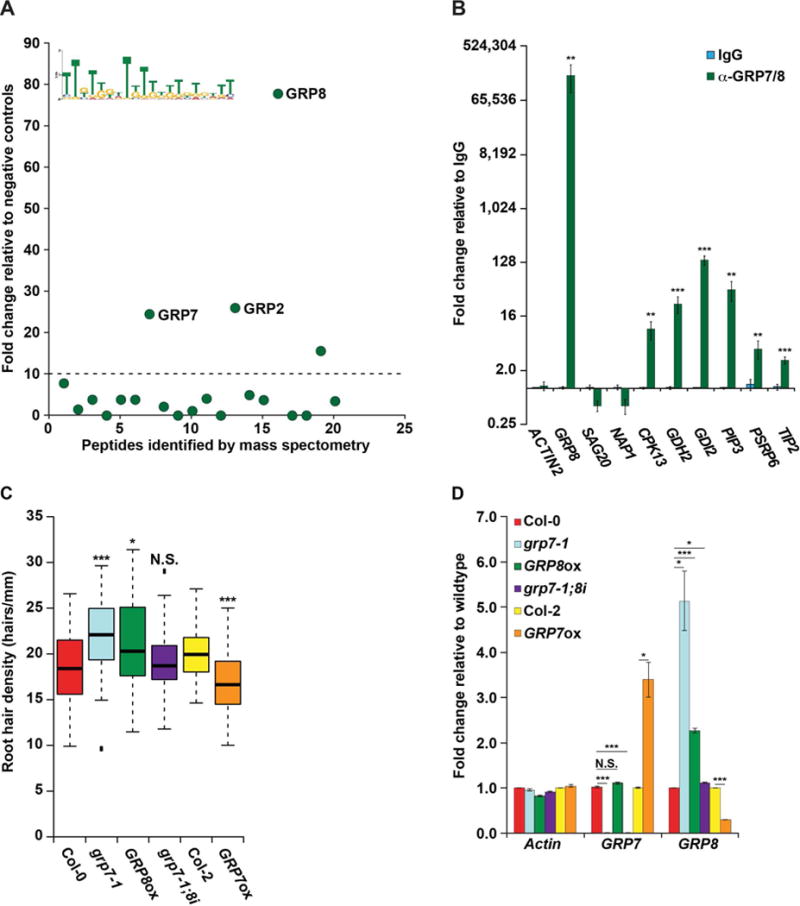
(A) RNA affinity chromatography followed by LC-MS was performed on whole root cell lysate using the MEME identified TG-rich motif as bait. Peptides above the dotted line have a more than 10-fold change and are candidate RBPs, with three GRPs denoted. (B) RIP-qPCR was performed on whole root lysate using rabbit IgG (blue bars) or rabbit serum raised against GRP7 and GRP8 (green bars) graphed as fold change relative to IgG. (C) Root hair cell density was measured in 8-day-old seedlings of WT or plants with decreased or increased GRP7 (grp7-1 or GRP7ox, respectively), increased GRP8 (GRP8ox), or decreased GRP7 with WT levels of GRP8 (grp7-1;8i), n > 50. * and *** denote p value < 0.05 and 0.001, respectively, while N.S. denotes p value > 0.05, Wilcoxon test. (D) RT-qPCR of root tissue from lines with altered GRP7 and/or GRP8 levels, graphed as fold change relative to WT (Col-0 or Col-2). For (B) and (D), *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Welch’s t-test. Error bars indicate SEM. See also Figures S1, S2, S4, S6, and S7.
