Fig. 3.
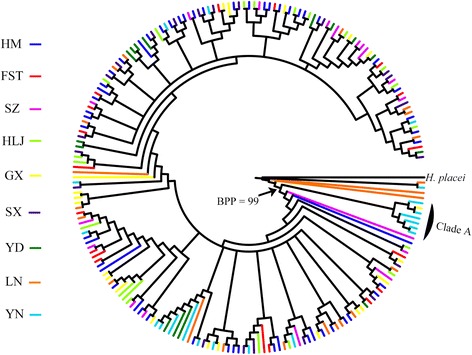
Phylogenetic tree resulting from Bayesian analysis (70 million generations) of 215 nad4 sequences of H. contortus (55 from HM, 18 from FST, 21 from SZ, 15 from YD, 21 from GX, 20 from YN, 23 from SX, 22 from LN, 20 from HLJ). The likelihood values of the 50% majority consensus tree was lnL = -3971.767. The average standard deviation of split frequencies was 0.007252 and all ESS values >200. The different coloured branches represent nad4 sequences from the different populations/sampling locations. Posterior probabilities were too low and not displayed in the tree. Abbreviations: FST, Farm Seven Team; GX, Guangxi; HLJ, Heilongjiang; HM, Helan Mountains; LN, Liaoning; SX, Shaanxi; SZ, Suizhou; YD, Yidu; YN, Yunnan
