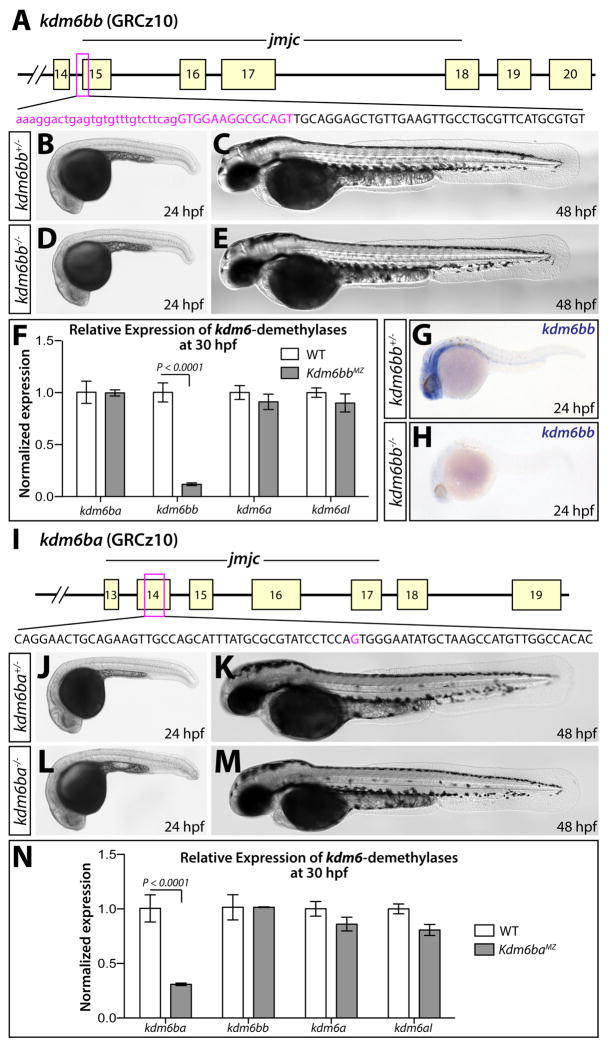
Figure 2. Generation and characterization of kdm6b loss of function alleles.
(A) Schematic showing the deleted sequence in the kdm6bb mutant allele recovered from CRISPR/Cas9 targeted mutagenesis (magenta bases). (B–E) Whole mount differential interference contrast (DIC) microscopy images of 24 (B and D) and 48 hpf (C and E) embryos. (F) Bar graph showing relative kdm6bb transcript levels between wildtype and kdm6bbMZ embryos at 30 hpf determined by qRT-PCR on cDNA from three separate pools of ten embryos each. (G, H) In situ hybridization for kdm6bb on control and kdm6bbMZ 24 hpf zebrafish embryos. (I) Schematic showing the CRISPR/Cas9-generated kdm6ba mutant allele. The deleted base is shown in magenta. (J–M) DIC images of control and kdm6baMZ embryos at 24 hpf (J and L) and 48 hpf (K and M). (N) Bar graphs comparing qRT-PCR-determined normalized expression levels of each H3K27me3 demethylase-encoding transcripts between 30 hpf wildtype and kdm6bbMZ embryos (three independent pools of ten embryos each). Error bars in the bar graphs (F, N) show one standard deviation. P-values indicate a significant difference determined by Student’s two-tailed t-tests after applying a Bonferroni correction for multiple comparisons.