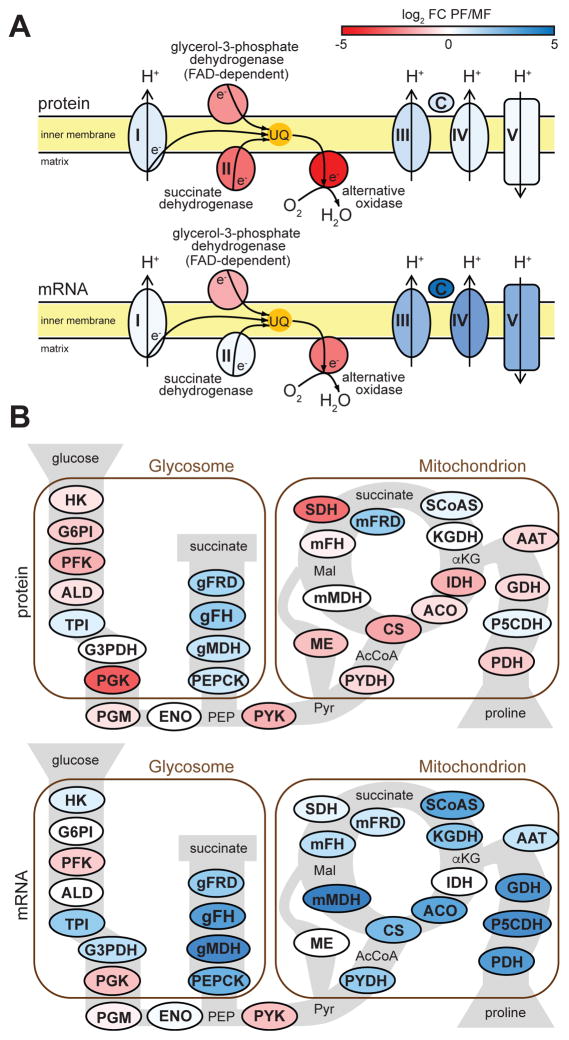
Figure 4. Changes in energy metabolism in metacyclics.
(A) Components of the mitochondrial respiratory chain (multisubunit complexes are represented by a single protein component in the figure): complex I, NADH-ubiquinone oxidoreductase (Tb927.5.450); glycerol-3-phosphate dehydrogenase (Tb927.11.7380); succinate dehydrogenase (Tb927.9.5960); alternative oxidase (Tb927.10.7090), complex III, rieske iron-sulfur protein (Tb927.9.14160); cytochrome c (Tb927.8.5120); complex IV, cytochrome oxidase subunit IV (Tb927.1.4100); complex V, ATP synthase alpha chain (Tb927.7.7420). (B) Glycolysis, TCA cycle and proline metabolism (multisubunit complexes are represented by a single protein component in the figure): HK, hexokinase (Tb927.10.2010); G6PI, glucose-6-phosphate isomerase (Tb927.1.3830); PFK, phosphofructokinase (Tb927.3.3270); ALD, aldolase (Tb927.10.5620); TPI, triosephosphate isomerase (Tb927.11.5520); G3PDH, glyceraldehyde 3-phosphate dehydrogenase (Tb927.6.4280); PGK, phosphoglycerate kinase (Tb927.1.700); PGM, phosphoglycerate mutase (Tb927.10.7930); ENO, enolase (Tb927.10.2890); PEPCK, phosphoenolpyruvate carboxykinase (Tb927.2.4210); gMDH, glycosomal malate dehydrogenase (Tb927.10.15410); gFH, glycosomal fumarate hydratase (Tb927.3.4500); gFRD, glycosomal fumarate reductase (Tb927.5.930); PYK, pyruvate kinase (Tb927.10.14140); PYDH, pyruvate dehydrogenase (Tb927.3.1790); CS, citrate synthase (Tb927.10.13430); ACO, aconitase (Tb927.10.14000); IDH, isocitrate dehydrogenase (Tb927.8.3690); KGDH, α-ketoglutarate dehydrogenase (Tb927.11.1450); SCoAS, succinyl-CoA synthetase (Tb927.3.2230); SDH, succinate dehydrogenase (Tb927.9.5960); mFRD, mitochondrial fumarate reductase (Tb927.5.940); mFH, mitochondrial fumarate hydratase (Tb927.11.5050); mMDH, mitochondrial malate dehydrogenase (Tb927.10.2560); ME, malic enzyme (Tb927.11.5450); PDH, proline dehydrogenase (Tb927.7.210); P5CDH, pyrroline-5 carboxylate dehydrogenase (Tb927.10.3210); GDH, glutamate dehydrogenase (Tb927.9.5900); AAT, alanine aminotransferase (Tb927.1.3950). The RNA-Seq fold-change values used here were obtained by the DNASTAR analysis (see Experimental Procedures).