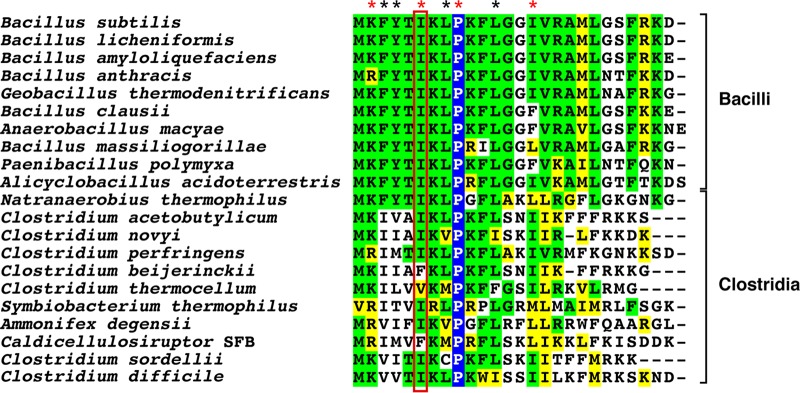
FIG 1 .
Sequence alignment of VM homologs in the Bacilli and Clostridia. Pro9 is the only residue that is completely conserved (boxed in blue with white font). This residue introduces a kink into the helical structure of VM (18) and is essential for it to specifically recognize positively curved membranes (19, 24). Conserved identical residues are boxed in green, and conserved similar residues are boxed in yellow. Sequences were derived from the work of Galperin et al. (7). Residues whose mutation to alanine causes severe defects in B. subtilis VM function (35) are indicated with an asterisk, while red asterisks highlight residues that have been shown or are presumed to activate the CmpA pathway in B. subtilis. K2A and I15A mutations prevent cortex production and activate the CmpA-dependent checkpoint pathway (26, 27), while I6A (highlighted by the red box) prevents binding to IVA and IVA encasement of the forespore (22) and thus presumably also activates CmpA.