Figure 2.
Single-Cell Transcriptome Analysis Distinguishes Various Cell Lineages Labeled by Lgr5 and Lgr6
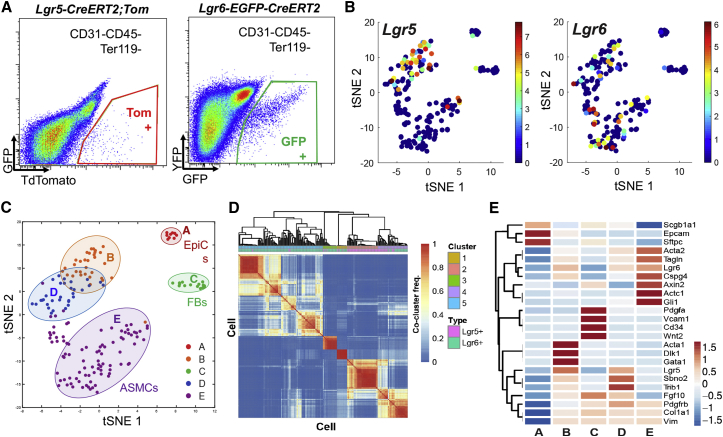
(A) Representative profile of FACS-sorted GFP+ populations for Lgr6-expressing cells from Lgr6-EGFP-CreERT2 mice (left) and Tom+ populations for Lgr5-expressing cells from Lgr5-CreERT2;R26-Tom mice post-induction (right) for single-cell RNA sequencing.
(B and C) T-stochastic neighbor embedding (tSNE) plot of 182 individual cells isolated in (A) (dots), where cells are either colored by the expression of Lgr5 (B, left) or Lgr6 (B, right; color bar, log2(TPM+1), left) or by their cluster assignment (C).
(D) Consensus clustering. Heatmap shows for each cell (rows and columns) and the frequency of times. Pairs of cells are clustered into the same cluster in 1,000 clustering applied to 1,000 random subsamples (color bar, 0, blue; 1, red). The matrix is hierarchically clustered (dendrogram, top). The final consensus cell clusters are marked by color code (A–E). Lgr5+ and Lgr6+ cells are marked in pink and green.
(E) Heatmap showing the relative average expression for selected marker genes across the 5 clusters.
See also Figure S2.