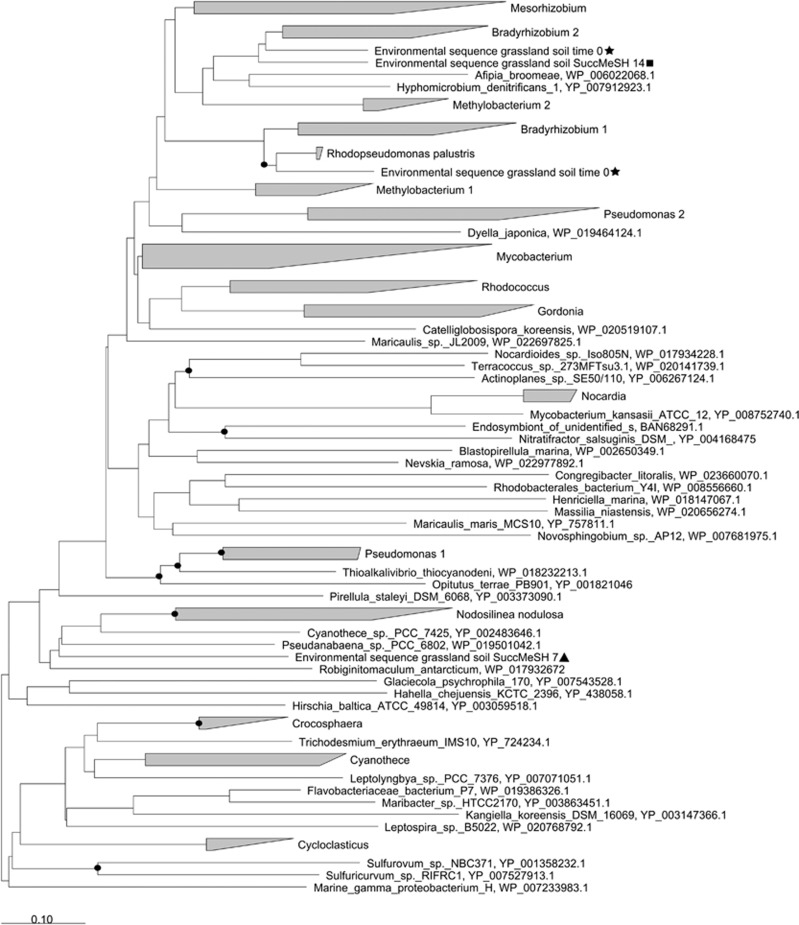
Figure 4.
Neighbour-joining phylogenetic tree of mddA including the environmental sequences from the assembled metagenomes of the grassland soil. Stars: sequences obtained from the metagenome of the grassland soil at time 0; Triangles: sequences obtained from the metagenome of the succinate plus MeSH enrichment after 7 days; Squares: sequences obtained from the metagenome of the succinate plus MeSH enrichment after 14 days. The accession numbers of the proteins encoded by the nucleotide sequences used in this tree are shown. Sequences with 57% identity to each other that are in bacteria of the same genus are shown in triangles; the size of the triangle reflects the number of sequences. Bar, 0.10 substitutions per nucleotide position. Bootstrap values ⩾70% (based on 1000 replicates) are represented with dots at branch points.