FIG 1.
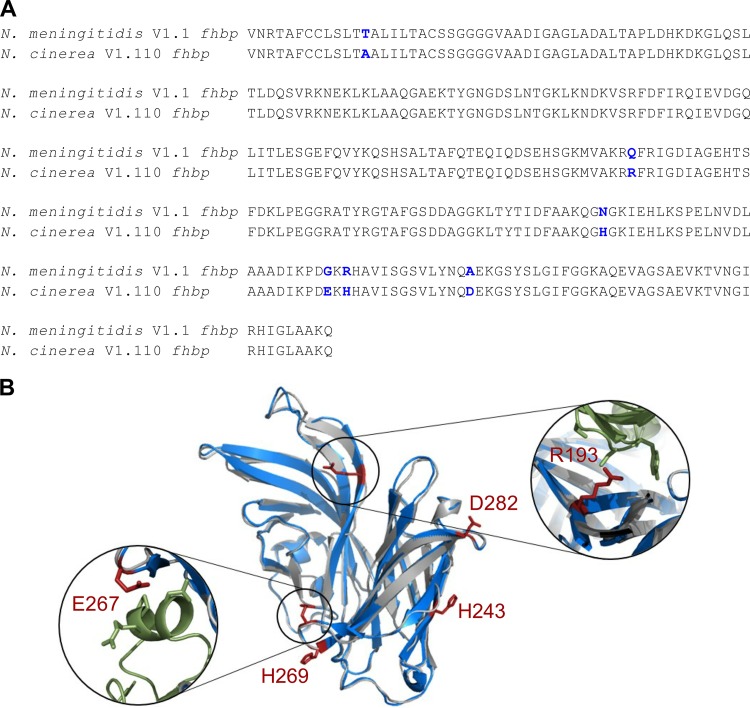
N. cinerea fHbp is predicted to be structurally similar to V1.1 fHbp. (A) Sequence alignment of N. meningitidis V1.1 fhbp and N. cinerea V1.110 fhbp. The blue residues show sequence differences. (B) Predicted structure of N. cinerea V1.110 fHbp (gray ribbon) generated with the I-TASSER server and V1.1 fHbp (blue ribbon [25]; PDB accession no. 2W80). The figure was drawn using Pymol. Residues in red (shown as ball-and-stick representations) are different in N. cinerea fHbp and V1.1 fHbp. Residues R193 and E267 (red) are in close proximity to the CFH (green ribbon) binding site.