Figure 6.
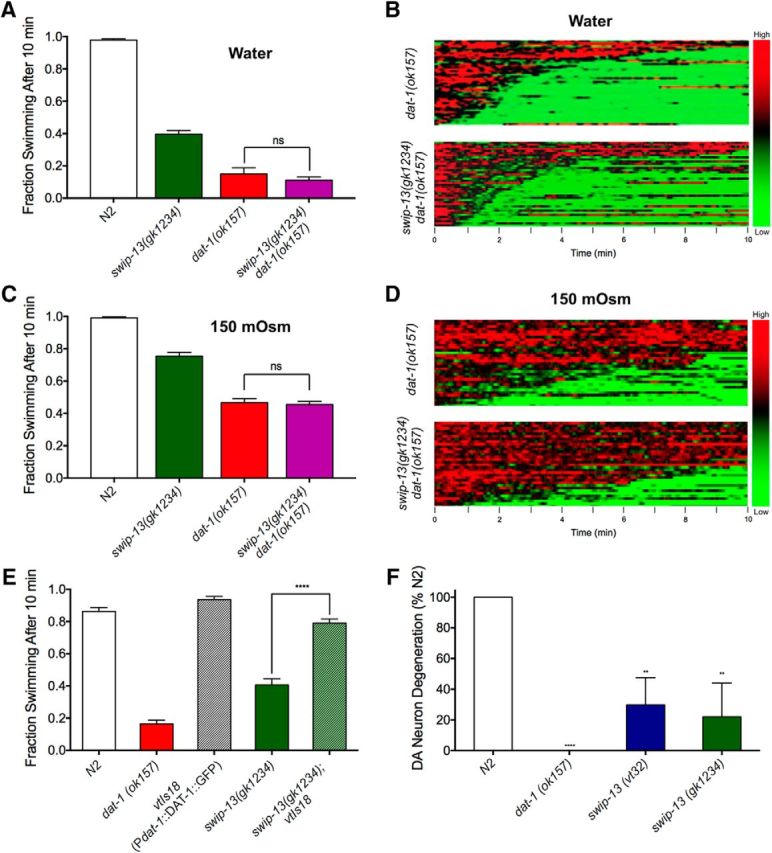
SWIP-13 regulates DAT-1 to control DA signaling. A, In water, dat-1(ok157) and swip-13(gk1234) dat-1(ok157) mutants display similar Swip behavior, with no significant difference between genotypes (t(92) = 1.11, p = 0.27, Bonferroni's post-tests). B, To increase the range in which we might see additivity between swip-13(gk1234) and dat-1(ok157), we performed Swip assays in 150 mOsm sucrose-supplemented water to suppress Swip. In this context, there is still no significant difference between dat-1(ok157) and swip-13(gk1234) dat-1(ok157) animals (t(92) = 0.44, p = 0.66, Bonferroni's post-tests). Data were analyzed using a one-way ANOVA with Bonferroni's post-tests comparing dat-1(ok157) to swip-13(gk1234) dat-1(ok157) in both A and B. C, D, Heat map analysis shows no enhancement of Swip in swip-13(gk1234) dat-1(ok157) animals compared with dat-1(ok157) mutants in either water (C) or 150 mOsm solution (D). E, A functional DAT-1::GFP-expressing transgene vtIs18 (Pdat-1::dat-1::gfp) rescues the paralysis of swip-13(gk1234) mutant animals. This is demonstrated by the significant difference between swip-13(gk1234) and swip-13(gk1234); vtIs18 animals (t(155) = 14.52, p = 0.27, Bonferroni's post-tests). Data were analyzed using a one-way ANOVA with Bonferroni's post-tests comparing swip-13(gk1234) to swip-13(gk1234); vtIs18. Significance was set at p < 0.05. ****p < 0.0001. F, Treatment of N2 animals with GFP-labeled DA neurons with the DA-neuron selective, DAT-1-dependent neurotoxin 6-OHDA leads to robust degeneration of DA neurons, as measured by loss of GFP-labeled CEP dendrites. dat-1(ok157) mutants are insensitive to this neurotoxin, shown as an absence of DA neuron degeneration. Both swip-13(vt32) and swip-13(gk1234) have reduced sensitivity to 6-OHDA compared with N2 animals [swip-13(v32): t(8) = 3.51, p < 0.01, Bonferroni's post-tests; swip-13(gk1234): t(8) = 3.9, p < 0.01, Bonferroni's post-tests]. Data were analyzed using a one-way ANOVA with Bonferroni's post-tests comparing all genotypes to N2. Significance was set at p < 0.05. **p < 0.01 ****p < 0.0001.
