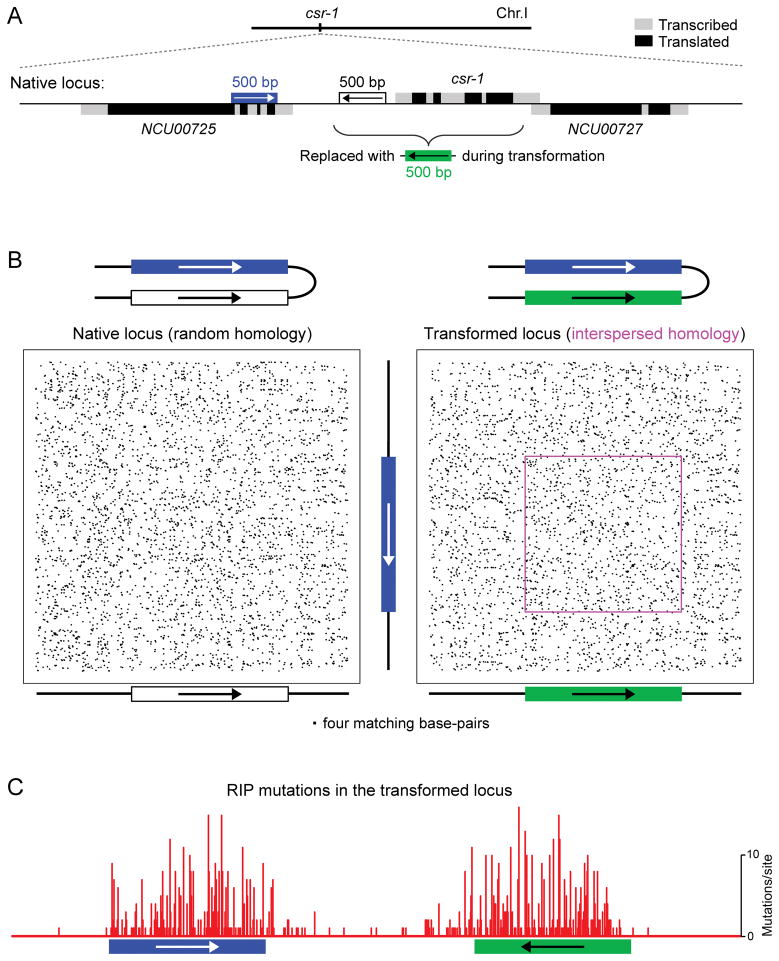
FIGURE 2.
Recognition of interspersed homology during RIP in N. crassa. This assay detects and quantifies the occurrence of RIP mutations in response to engineered DNA repeats. Instances of DNA homology are created between two short segments of chromosomal DNA, one of which is normally represented by an endogenous sequence, while the sequence and orientation of the other segment can be manipulated as desired. In this situation, the number of RIP mutations provides a very sensitive readout of DNA homology perceived by the recombination-independent mechanism of repeat recognition for RIP. (A) Weak interspersed homology is formed between the endogenous 500-bp segment (blue) and a synthetic DNA segment (green) integrated at a nearby position as the replacement of the cyclosporin-resistant-1 (csr-1) gene. This particular pattern involves 4-bp units of homology spaced with the periodicity of 11 base-pairs and exists between “repeat units” in the inverted orientation. (B) Pairwise sequence comparisons showing all matches of 4 base pairs long. Two situations are presented: random homology (left panel) and interspersed homology (right panel). No cryptic homology can be seen except the intended pattern of weak interspersed homology (magenta box). (C) The occurrence of mutations induced by weak interspersed homology. Seventy progeny spores from the “XKO” cross (70), which had been previously found to contain at least one RIP mutation, were reanalyzed by sequencing of additional 255 base pairs in the “left” flank of the construct (corresponding to the single-copy coding/translated sequence of NCU00725).