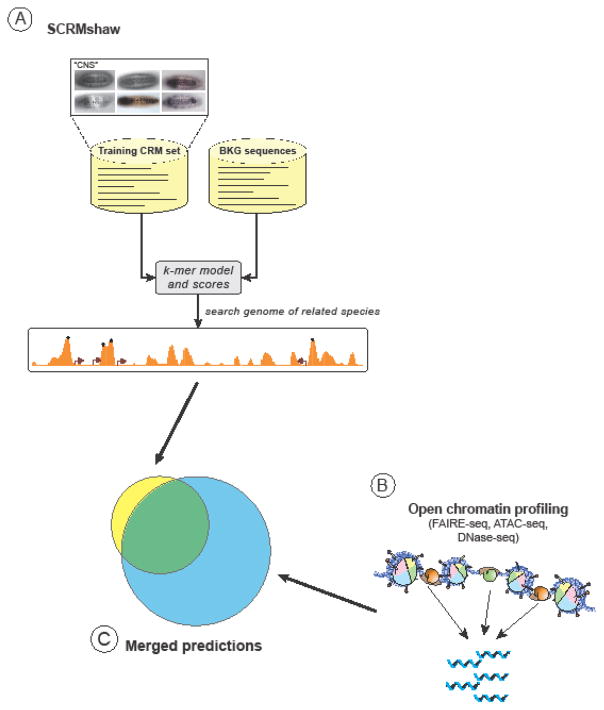
Figure 4. CRM discovery across distantly-related species.
(A) The SCRMshaw method [46–48] can be used to identify CRMs in diverged insect species. Training data composed of the sequences of known Drosophila melanogaster CRMs with a common function (e.g., expression in the central nervous system (“CNS”), top of figure) are compared to randomly selected non-coding sequences (“BKG” or “background” sequences). A scoring model (“k-mer model and scores”) is then generated that can be used to search the genome of another insect species to predict CRMs. The method has been shown to be effective at least through 345 million years of holometabolous insect evolution. (C) To help reduce the roughly 25% false-positive prediction rate, SCRMshaw predictions can be merged with putative CRM regions predicted from open chromatin profiling methods (B) such as FAIRE-seq [58] or ATAC-seq [64].