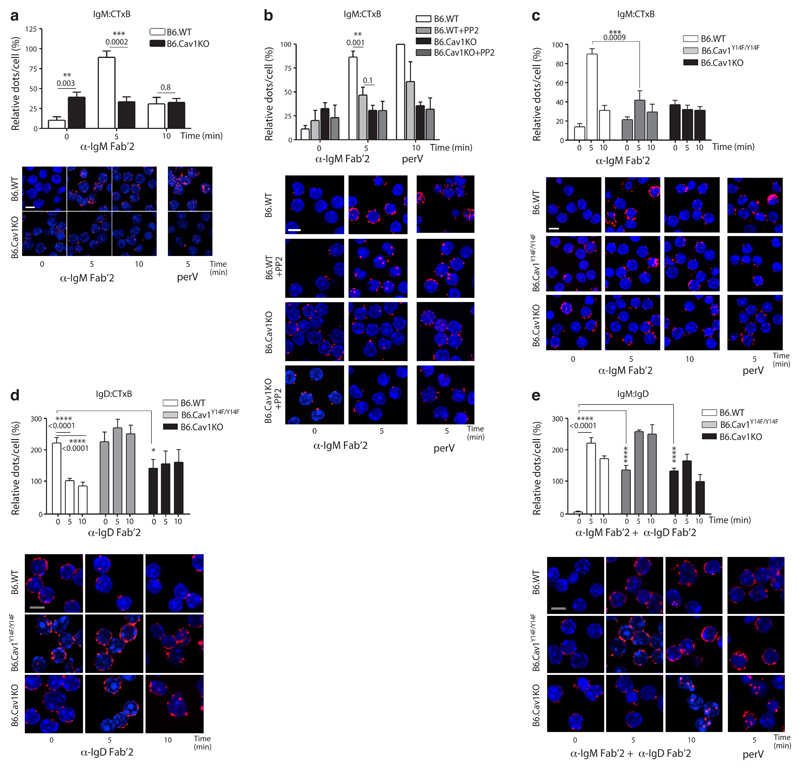
Figure 1. Cav1 regulates IgM- and IgD-BCR nanoclustering.
(a) Purified splenic B cells were stimulated with 10 μg/ml of anti-IgM Fab‘2 for the indicated times or left unstimulated. PLA analyzes the proximity between the IgM-BCR and GM1-ganglioside. PLA signals (red dots per cell) were counted and normalized to the PLA signals of B6.WT cells stimulated for 5 min with 0.5 mM pervanadate (perV) for each individual experiment. Representative microscope images show DAPI stained nuclei (in blue) and PLA signals (in red). (b) Purified splenic B cells were pretreated with 10 μM PP2 or vehicle for 60 min before being stimulated and analyzed as in a. (c) Cells were stimulated and analyzed as in a. ANOVA P=0.004. (d) Purified splenic B cells were stimulated with 10 μg/ml of anti-IgD Fab‘2 for the indicated times or left unstimulated. PLA analyzes the proximity between the IgD-BCR and GM1-ganglioside. ANOVA P=0.05. (e) Purified splenic B cells were stimulated with 10 μg/ml of anti-IgM Fab‘2 and anti-IgD Fab‘2 for the indicated times. PLA analyzes the proximity between IgM-BCR and IgD-BCR. ANOVA P<0.0001.
In all panels, quantified data of at least three experiments were pooled and statistical analysis performed using unpaired Student’s t test when two conditions were compared. ANOVA test was used to compare B6.Cav1KO and B6.Cav1Y14F to B6.WT samples in unstimulated conditions followed by Dunnett's multiple comparisons. When significant, P values are indicated. Means ± SEM are shown. At least 300 cells were quantified per experimental condition and experiment. Size bar 5 μm.