Figure 1. Ectopic expression of miR-K6-5p promotes endothelial cells invasion and angiogenesis in vitro.
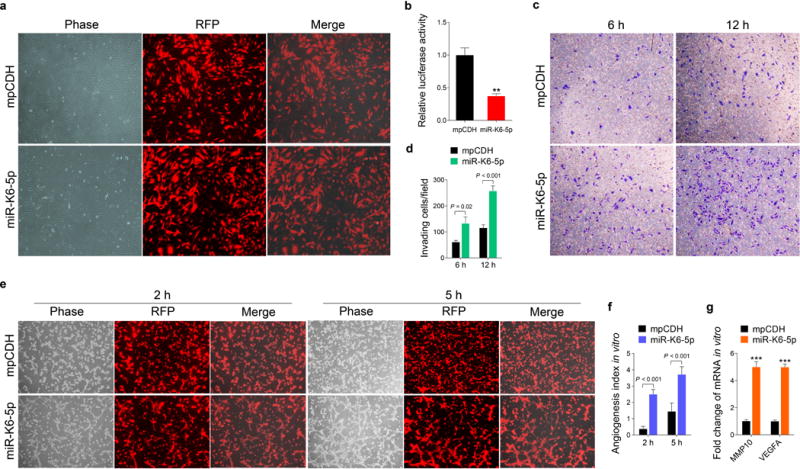
(a). HUVECs were transduced with lentivirus empty vector (mpCDH; top) and lentivirus-miR-K6-5p (miR-K6-5p; bottom), and representative images were taken under the light microscope (Phase) and fluorescent microscope (RFP) (Original magnification, ×100).
(b). Luciferase activity was detected in lentivirus empty vector (mpCDH) or lentivirus-miR-K6-5p (miR-K6-5p) transduced HUVECs followed by transfection with the pGL3-Control (Control) or the pGL3-miR-K6-5p sensor reporter (Sensor). ** P < 0.01 for Student’s t-test versus mpCDH group.
(c). Matrigel invasion assay with HUVECs transduced with lentivirus empty vector (mpCDH) or lentivirus-miR-K6-5p (miR-K6-5p). The representative images were captured at 6 h and 12 h post seeding (original magnification, ×100).
(d). The quantified results of Matrigel invasion assay in (c). The quantified results represent the mean ± SD. Three independent experiments, each with five technical replicates, were performed.
(e). Microtubule formation assay for HUVECs transduced with lentivirus empty vector (mpCDH; top) or lentivirus-miR-K6-5p (miR-K6-5p; bottom). The representative images were captured under the light microscope (Phase) and fluorescent microscope (RFP) at 2 and 5 h post seeding (original magnification, ×100).
(f). The quantified results of microtubule formation assay in (e). The quantified results represent the mean ± SD. Three independent experiments, each with five technical replicates, were performed.
(g). The mRNA expression levels of MMP10 and VEGFA in HUVECs treated as in (a) were determined by RT-qPCR. The quantified results represent the mean ± SD. Three independent experiments, each with four technical replicates, were performed. *** P < 0.001 for Student’s t-test versus mpCDH group.
