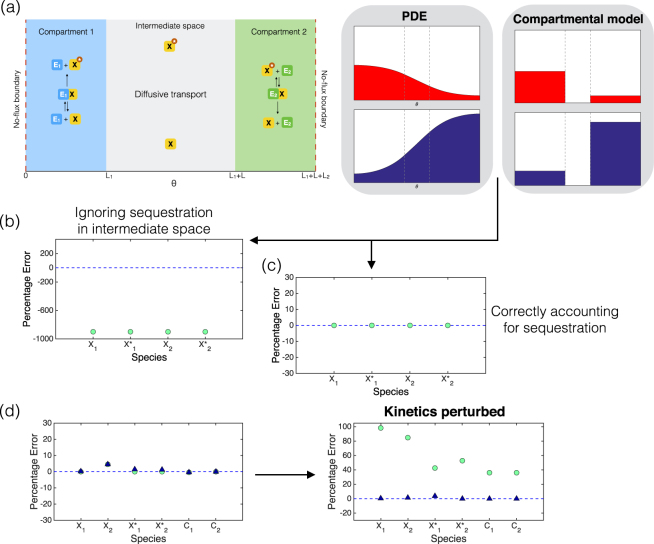
Figure 1.
Compartmentalized single modification cycle. (a) Schematic for the compartmentalized pathway. The PDE description yields steady state concentration profiles of X and X*. The compartmental ODE gives steady state compartmental concentrations of X and X*. We examine the error between the PDE compartmental averages and the ODE steady state. (b) Percentage error for each species, between the steady state compartmental averages of the PDE model, and the steady state of a compartmental model using the same total amount of substrate as the PDE model. We consider a system with two compartments, each occupying 5 percent of the whole domain. (c) In the mass action regime, if the total amount of substrate in the compartments is exactly accounted for, transport parameters can be computed, which result in an exact match. (d) In the non-mass action regime, an exact fit cannot be obtained, even if the total amount of substrate in the compartments is accounted for. Errors are shown for both the basic compartmental model calibrated with the exact total amount of substrate in the compartments under basal conditions - green circles, and the compartmental model with modified conservation to account for amount of substrate in the intermediate space – blue triangles. For the basal kinetics, both cases match the PDE closely. When the kinetic parameter is changed (phosphatase concentration in compartment 2 increases), the modified model (blue triangles) continues to give a good fit, while the basic compartmental model with calibration under basal conditions (green circles) produces large errors.