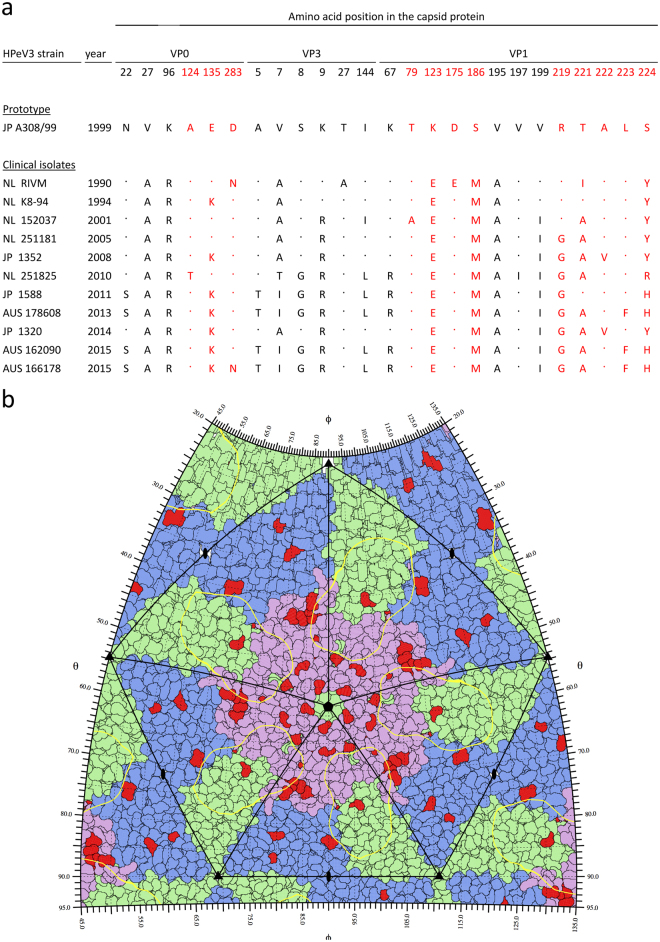
Figure 2.
Amino acid variation in the capsid-encoding region of a subset of HPeV3 isolates. (a) Alignment showing the positions of variable amino acid residues. Capsid surface-exposed residues are highlighted in red. (b) HPeV3 roadmap of capsid exterior. Five full asymmetric units are shown; each formed of one copy of capsid proteins VP0 (blue), VP3 (green) and VP1 (pink). The AT12-015 Fab footprint (yellow contour) is mapped on surface residues differing between the strains (red), in comparison to the prototype A308/99. Residue-labeled close-up roadmaps for each strain are showed in Supplementary Information 1.