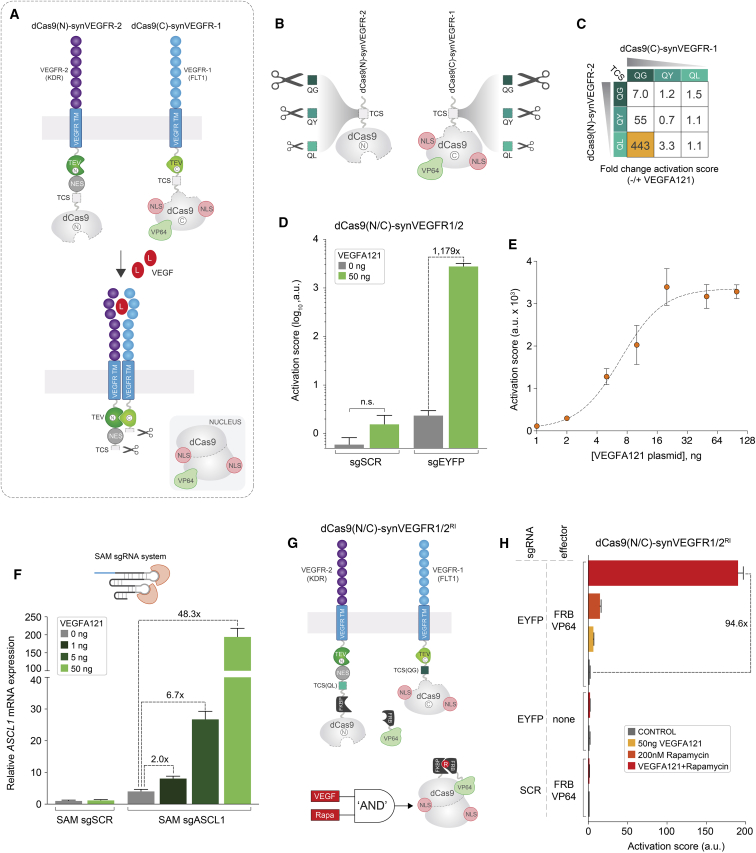
Figure 2.
Construction and Optimization of a Prototype Chimeric dCas9-synRTK
(A) Design principles underlying the generation of a VEGF-responsive dCas9-synRTK.
(B and C) Optimization of chimeric dCas9(N/C)-synVEGFR1/2 performance by fine-tuning coordinated signal release efficiency. Three TCS variants (QG, QY, QL) of decreasing strength were sequentially grafted on both the dCas9(N)-synVEGFR2 and dCas9(C)-synVEGFR1 (B), and the competency of all possible combinations to drive EYFP expression was tested in the presence or absence of VEGFA121 agonist (C) (see Figure S3C).
(D) Quantification of EYFP activation score for the top candidate from (C) complemented either with control sgRNA (sgSCR) or targeting sgRNA (sgEYFP), in the presence or absence of VEGFA121 agonist (n = 3 from one experiment, mean ± SD; a.u., arbitrary units; GraphPad Prism unpaired two-sided t test with Welch’s correction, not significant [n.s.] p > 0.05).
(E) Dose-response curve for the dCas9(N/C)-synVEGFR1/2 variant in (D) at increasing concentrations of VEGFA121 plasmid. Each data point represents EYFP activation score from 3 biological replicates (mean ± SD, a.u. arbitrary units; curve was fitted using a non-linear variable slope [four parameters] function in GraphPad Prism).
(F) Programmed activation of endogenous gene expression by dCas9(N/C)-synVEGFR1/2 in the presence of control SAM sgRNA (SAM sgSCR) or a pool of ASCL1-targeting SAM sgRNAs (SAM sgASCL1), and increasing concentrations of VEGFA121 plasmid (n = 3 biological replicates [×3 technical replicates], mean ± SD).
(G) Schematic representation of AND gate control for dCas9(N/C)-synVEGFR1/2RI activation.
(H) Analysis of EYFP induction by dCas9(N/C)-synVEGFR1/2RI in the absence of any inducer (CONTROL), in the presence of VEGFA121 plasmid alone, rapamycin alone, and combined delivery of both inducers. A control sgRNA (sgRNA SCR) and reactions lacking the FRB-VP64 effector construct were used to establish baseline activation.
In all cases the EYFP activation score was calculated from three biological replicates (n = 3 from one experiment, mean ± SD; a.u., arbitrary units; sgSCR, scramble sgRNA control). See also Figure S3.