Figure 3.
Programmed Control of Gene Expression with Chimeric dCas9-synGPCRs
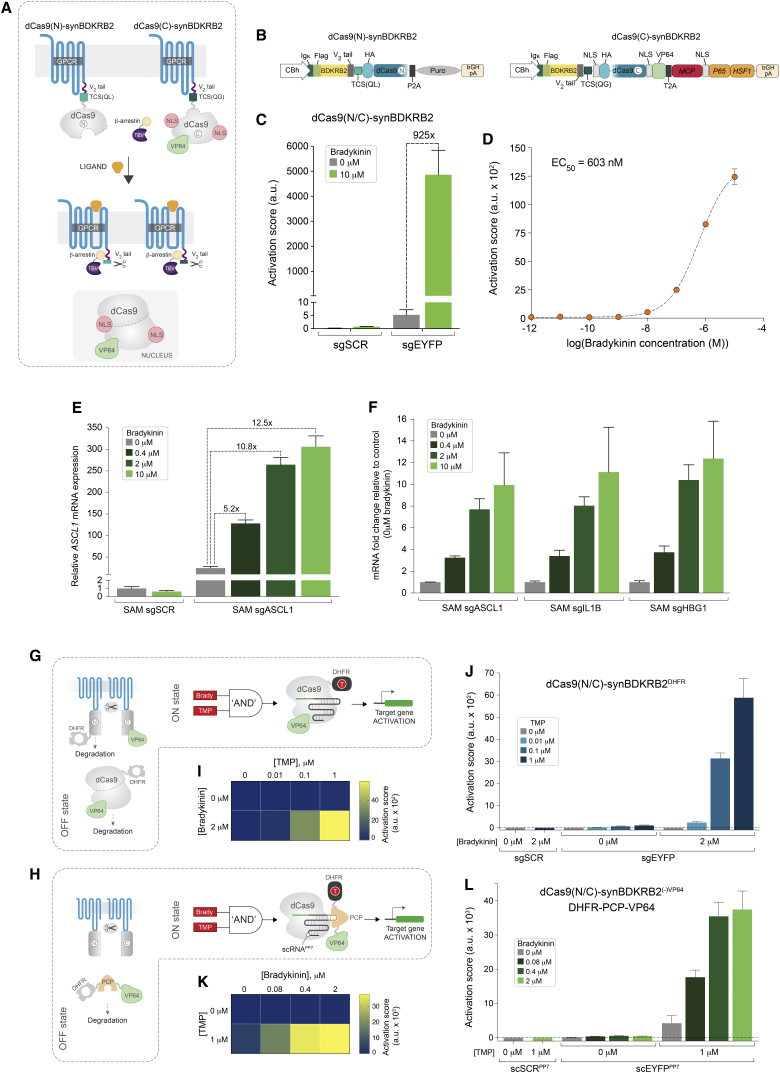
(A) Schematic representation of dCas9-synGPCR design concept.
(B) Diagrams of the prototype dCas9(N/C)-synBDKRB2 receptor constructs.
(C) Quantification of EYFP activation score following bradykinin-mediated induction of dCas9(N/C)-synBDKRB2 in HTLA cells constitutively expressing β-arrestin2-TEV fusion protein (n = 3 biological replicates from one experiment, mean ± SD, a.u. arbitrary units; sgSCR, scramble sgRNA).
(D) Dose-response curve for dCas9(N/C)-synBDKRB2 complemented with sgEYFP guide RNA at increasing concentrations of bradykinin (EC50 = half-maximal effective concentration; each data point represents EYFP activation score from 3 biological replicates, mean ± SD, a.u. arbitrary units; curve fitted using a non-linear variable slope [four parameters] function in GraphPad Prism).
(E) Induced expression of endogenous ASCL1 gene by dCas9(N/C)-synBDKRB2 in HTLA cells. Graph shows ASCL1 mRNA expression levels using a pool of ASCL1 sgRNAs (SAM sgASCL1) relative to control sgRNA (SAM sgSCR) at increasing concentrations of bradykinin.
(F) dCas9(N/C)-synBDKRB2-mediated dose-dependent activation of three target genes (ASCL1, IL1B, HBG1) at increasing agonist concentrations (0.4, 2, 10 μM bradykinin), displayed as fold change relative to no-agonist conditions (0 μM bradykinin). Values in (E) and (F) were calculated from n = 3 biological replicates (×3 technical replicates), mean ± SD.
(G and H) Schematic representation of two “AND” gate ON-switch strategies for dCas9(N/C)-synGPCR activation, whereby the DHFR destabilization domain is fused directly to either the dCas9(N) fragment (G) or the PCP-VP64 effector protein (H).
(I and J) Heatmap (I) and quantification (J) of reporter activation in the absence or presence of bradykinin and increasing concentrations of TMP, following the strategy described in (G).
(K and L) Heatmap (K) and quantification (L) of reporter activation in the absence or presence of TMP and increasing concentrations of bradykinin, following the strategy described in (H).
In all cases EYFP activation score was calculated from three biological replicates (n = 3 from one experiment, mean ± SD; a.u., arbitrary units; sgSCR, scramble sgRNA; scSCRPP7, scramble PP7-aptamer scaffold guide RNA; scEYFPPP7, EYFP-targeting PP7-aptamer scaffold guide RNA). See also Figure S5.