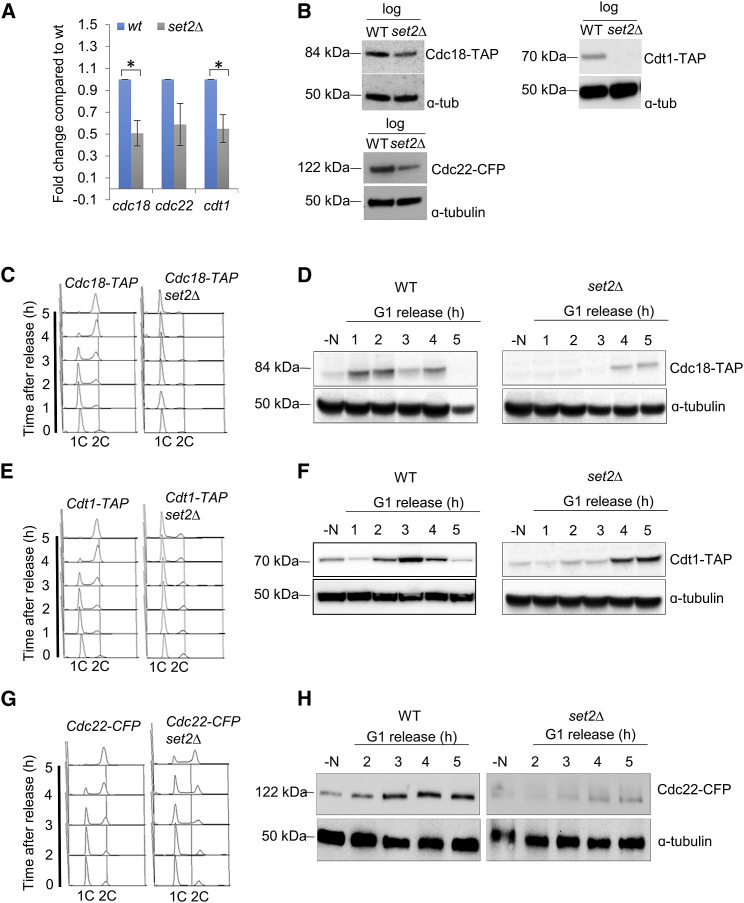
Figure 4.
Replication Factors Are Misregulated in set2Δ Cells
(A) Transcript levels of cdc18+, cdc22+, and cdt1+ were established in exponentially growing wild-type and set2Δ cells. RNA was extracted with Qiagen RNeasy kit and relative transcript levels of cdc18+, cdc22+, and cdt1+ were established by RT-qPCR. Error bars represent SD of three biological repeats. The asterisk (∗) represents significant difference compared with wild-type and set2Δ.
(B) Cdc18, Cdt1, or Cdc22 protein levels of exponentially growing wild-type and set2Δ cells. Cell extracts were prepared from vegetative cells and processed for western blot. Immunoblots of total cell lysates were probed with PAP or GFP antibody. α-Tubulin is shown as a loading control.
(C) cdc18-TAP or cdc18-TAP set2Δ cells were arrested in G1 by nitrogen starvation and released, and samples were taken at time points indicated and subjected to FACS analysis.
(D) In parallel, cdc18-TAP and cdc18-TAP set2Δ cells were processed for western blotting at indicated times.
(E) cdt1-TAP or cdt1-TAP set2Δ cells were arrested in G1 by nitrogen starvation, released, and samples taken at time points indicated and subjected to FACS analysis.
(F) In parallel, cdt1-TAP and cdt1-TAP set2Δ cells were processed for western blotting at indicated times.
(G) A similar experiment to that described in (C) was carried out using cdc22-CFP or cdc22-CFP set2Δ cells.
(H) A similar experiment to that described in (D) was carried out using cdc22-CFP or cdc22-CFP set2Δ cells. Immunoblots of total cell extracts were probed with GFP antibody. α-Tubulin is shown as a loading control.