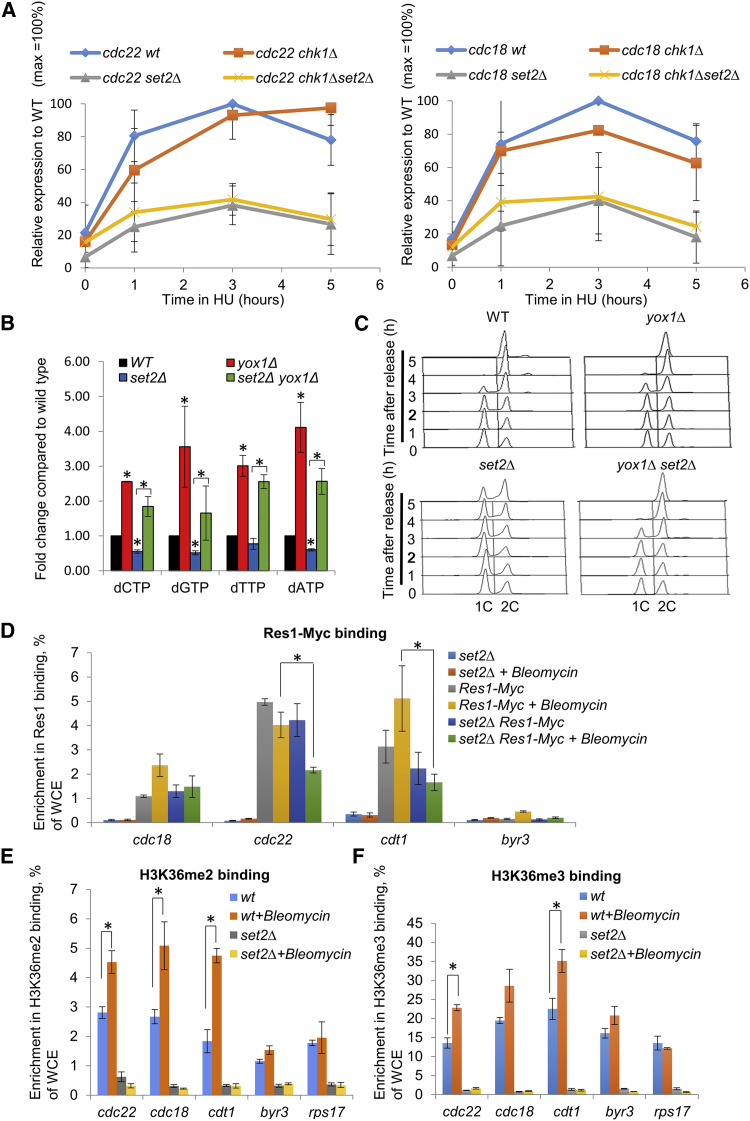
Figure 7.
Set2 Facilitates Efficient Binding of MBF Components in Response to Genotoxic Stress
(A) Set2 has a direct function in regulating MBF activity in response to replication stress. Left panel: quantification of cdc22+ transcript levels in wild-type, chk1Δ, set2Δ, and set2Δ chk1Δ cells following 5-hr treatment with 10 mM HU. Data represent mean of three experiments with independently derived RNA. Right panel: quantification of cdc18+ transcript levels in wild-type, chk1Δ, set2Δ, and set2Δ chk1Δ cells following 5-hr treatment with 10 mM HU. Data represent mean of three experiments with independently derived RNA.
(B) dNTP levels were measured in exponential growth wild-type, yox1Δ, set2Δ, and set2Δ yox1Δ strains. Means ± SEs of three experiments are shown. The asterisk (∗) represents significant difference compared with WT and set2Δ, WT and yox1Δ, set2Δ and set2Δyox1Δ.
(C) Removal of MBF inhibitor Yox1 speeds up S-phase progression in set2Δ cells. Flow cytometry analysis of the indicated strains after release from G1 arrest into EMM+N at 32°C.
(D) Levels of Res1-Myc binding at cdc18+, cdc22+, cdt1+, and byr3+ (negative control) promoters were shown in the presence or absence of DNA damage. Cells of logarithmically growing cultures (YES medium) of set2Δ, res1-Myc, set2Δ res1-Myc strains were collected for ChIP before and 30 min after treatment with 5 μg/mL bleomycin. Data represent the mean of three experiments with independently derived RNA and error bars (±SE) are shown. The asterisk (∗) represents significant difference compared with wild-type (p < 0.05, t test).
(E) Set2-dependent di-methylation of H3K36 at promoters is induced upon genotoxic stress. Levels of H3K36me3 at cdc18+, cdc22+, cdt1+, byr3+ (negative control), and rps17+ (negative control) promoters were presented in wild-type or set2Δ strains without and with DNA damage treatment. Cells of logarithmically growing wild-type and set2Δ cultures were collected for ChIP before and 30 min after treatment with 5 μg/mL bleomycin. Data represent the mean of three experiments with independently derived RNA and error bars (±SE) are shown. The asterisk (∗) represents significant difference compared with wild-type (p < 0.05, t test).
(F) Set2-dependent tri-methylation of H3K36 at promoters is induced upon genotoxic stress. Levels of H3K36me3 at cdc18+, cdc22+, cdt1+, byr3+ (negative control), and rps17+ (negative control) promoters were presented in wild-type or set2Δ strains without and with DNA damage treatment. Cells of logarithmically growing wild-type and set2Δ cultures were collected for ChIP before and 30 min after treatment with 5 μg/mL bleomycin. Data represent the mean of three experiments with independently derived RNA and error bars (±SE) are shown. The asterisk (∗) represents significant difference compared with wild-type (p < 0.05, t test).