Fig. 2.
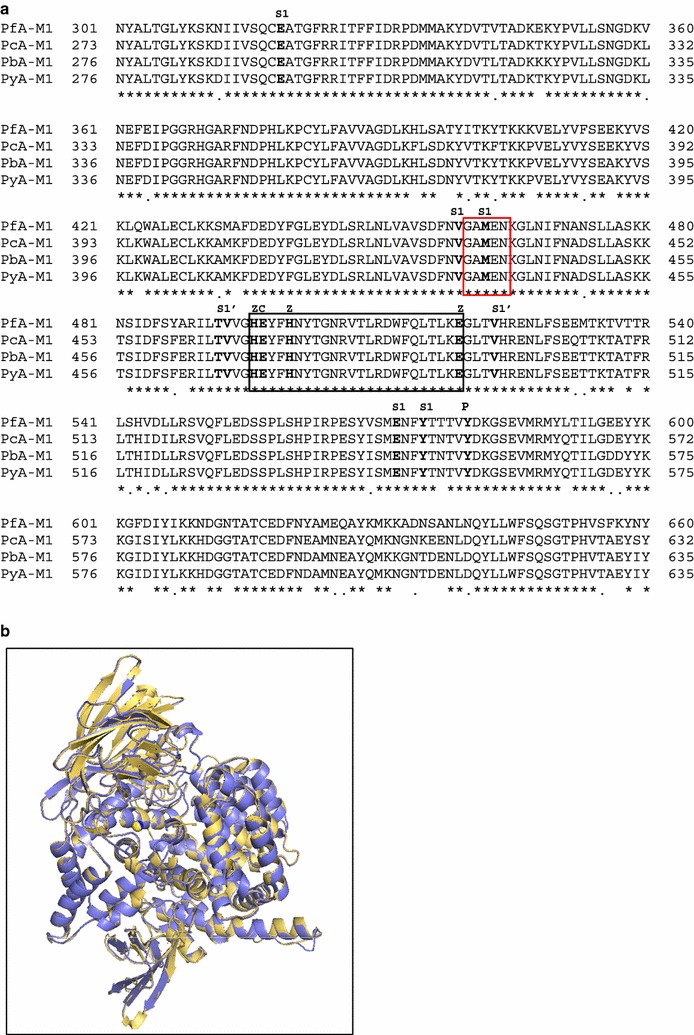
Comparison of M1 aminopeptidases from P. falciparum, P. c. chabaudi, P. berghei and P. yoelii. a Aminopeptidase M1 sequences from P. falciparum (O96935, PfA-M1), P. c. chabaudi (XP_745040.2, PcA-M1), P. berghei (XP_680130.1, PbA-M1) and P. yoelii (XP_729336.1, PyA-M1) were used for the multi alignment corresponding to domain 301–660 of the native PfA-M1 sequence. The active site HExxHx18E motif and the conserved GAMEN sequence are boxed in black and red, respectively. Identical (*) and conserved (.) amino acids between the four sequences are indicated. Key amino acids involved in S1 and S1′ subsites, in the ligation to zinc ion (Z), required for catalytic activity (C) and acting as putative proton donor (P) are indicated above alignment. b The 3D molecular model of PcA-M1 (blue) was compared to the 3D structure of PfA-M1 (PDB: 4J3B, yellow). Structures superposition was prepared using the PyMOL molecular graphics system
