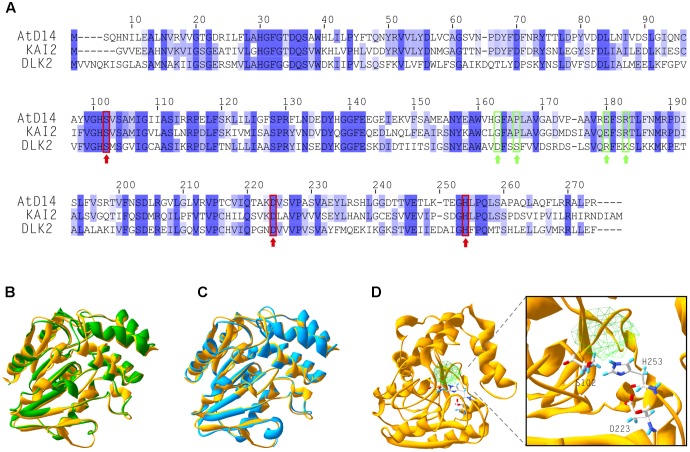
FIGURE 1.
DLK2 shares high structural similarity with KAI2 and AtD14. (A) Alignment of amino acid sequences of AtD14, KAI2 and DLK2 showing that DLK2 shares 40 and 42% identity at the protein level with KAI2 and AtD14, respectively. Dark blue color shows identity in all three proteins; light blue coloring shows identity in two of the three proteins. The amino acids of the catalytic triad are marked with red rectangles and arrows. The residues required for the physical interaction of AtD14 with MAX2 are marked with green rectangles and arrows. (B–D) Crystallized tertiary structures of AtD14 (B, green; PDB code 4IH4), KAI2 (C, blue; PDB code 4IH1) and predicted structure of DLK2 (D, yellow; (I-TASSER server; http://zhanglab.ccmb.med.umich.edu/I-TASSER/). (B,C) The predicted structure of DLK2 (yellow) is overlaid on those of AtD14 and KAI2, respectively, using Swiss-PdbViewer (Guex and Peitsch, 1997). An expanded view of the catalytic triad residues of DLK2 (Ser-102, Asp-223 and His-253) and the predicted cavity are shown in (D).