Abstract
We provide simple protocols for generating digital volume data with the high-resolution episcopic microscopy (HREM) method. HREM is capable of imaging organic materials with volumes up to 5 x 5 x 7 mm3 in typical numeric resolutions between 1 x 1 x 1 and 5 x 5 x 5 µm3. Specimens are embedded in methacrylate resin and sectioned on a microtome. After each section an image of the block surface is captured with a digital video camera that sits on the phototube connected to the compound microscope head. The optical axis passes through a green fluorescent protein (GFP) filter cube and is aligned with a position, at which the bock holder arm comes to rest after each section. In this way, a series of inherently aligned digital images, displaying subsequent block surfaces are produced. Loading such an image series in three-dimensional (3D) visualization software facilitates the immediate conversion to digital volume data, which permit virtual sectioning in various orthogonal and oblique planes and the creation of volume and surface rendered computer models. We present three simple, tissue specific protocols for processing various groups of organic specimens, including mouse, chick, quail, frog and zebra fish embryos, human biopsy material, uncoated paper and skin replacement material.
Keywords: Developmental Biology, Issue 125, Episcopic, imaging, 3D, modeling, embryo, biopsy, human, mouse, chick, zebra fish, frog
Introduction
Structural analysis of organic and anorganic materials is the first step in understanding their physical properties and function. The basis for such analysis often is two-dimensional (2D) information gained by careful observation of histological sections, with a variety of simple and sophisticated imaging methods that extract details of tissue architecture, cell morphology and topology, molecular composition and biomechanical properties1,2,3. However, 2D information is not suited for researching spatially complex arrangements. Hence a growing number of in vivo and ex vivo methods that permit the generation of digital volume data were established in the last decades4 and many more are under development.
The methodical principle of most volume data generation methods is the generation of virtual stacks of digital images displaying sections gained by virtual or physical sectioning of an object. If the section images are aligned properly, this creates a volume, which can be re-sectioned in virtual section planes, or used for creating 3D surface and volume rendered models. Popular techniques for visualizing humans and larger biological specimens are magnetic resonance tomography (MRT), computed tomography (CT), positron emission tomography (PET) and single-photon emission computed tomography (SPECT). Small specimens are usually visualized by using micro-magnetic resonance imaging (µMRI), optical projection tomography (OPT), optical coherence tomography (OCT), photoacoustic tomography (PAT), histological sectioning based methods, confocal microscopy, and electron tomography5,6,7,8,9,10,11,12,13,14,15,16,17.
A relatively new volume data generation technique, which produces digital data of small specimens and histological tissue samples is the HREM method, which was developed in close co-operation with Tim Mohun18,19. It is a simple microscope based technique, which generates digital volume data from resin embedded material that is sectioned on a microtome. The data facilitate detailed analysis of tissue architecture and cell distributions as well as metric analysis of small features on an intermediate light microscopic level.
HREM produces stacks of inherently aligned digital images that appear as if captured from eosin stained histological sections. Tissue contrast and data resolution in respect to the field of view exceed that of data produced with µCT, µMRI, and OPT, but are lower than that achievable with confocal, light sheet and electron microscopy20. However, in contrast to the latter, HREM is capable of visualizing specimens with relative large volumes of up to 5 x 5 x 7 mm3 in histological quality. A number of recent studies provide detailed characterizations and comparisons of advantages and disadvantages of the single imaging techniques and, for the sake of objectivity, we refer to those for further information regarding their limitations and potential fields of applications4,21,22,23,24.
This study focuses on the HREM imaging method and aims to provide very simple protocols for generating HREM data of a broad spectrum of organic materials, as well as examples of their application. The workflow for creating HREM data is simple and applies to all materials that can embed in methacrylate resin (Figure 1). Yet there are tissue specific differences in sample preparation, that need to be considered. We therefore provide three standard protocols for preparing various samples. Embedding and data generation protocol steps are identical for all of them.
Protocol
All procedures were performed in accordance with ethical guidelines at the Medical University of Vienna.
1. Sample Preparation
- Preparing embryos and embryo tissues (up to 5 x 5 x 5 mm3 )
- Fix the embryos or parts of the embryos in 4% PFA/PBS or Bouin's fixative at 4 °C for at least 16 - 24 h under constant rocking. NOTE: Embryos of several species and of various developmental stages can be used, such as 48-h zebrafish, 19-h chick and quail, and also prenatal mouse embryos, which were used here (see Representative Results). The specimens were harvested and transferred into PBS at room temperature for staging, before they were put into fixative. If necessary, trim the specimens with micro-scissors or scalpels prior to fixation to fit into the embedding molds.
- Remove the fixative and wash the embryo tissue in PBS at 4 °C under constant rocking for 24 h (2-3 changes).
- Dehydrate the samples in 70%, 80%, 90%, 96% ethanol at 4 °C for 2 - 3 h each. Place tubes containing the samples on a rotator. NOTE: For small samples (< 2 x 2 x 2 mm3), the time a sample rests in each ethanol may be reduced to 60 min. For large samples (> 5 x 5 x 5 mm3), it may be extended to 4 h.
- Perform the following infiltration steps under a fume hood wearing protective gloves.
- Prepare the infiltration solution by adding 1.25 g benzoyl peroxide (plasticized catalyst) and 0.4 g eosin to 100 mL of solution A (for more details, see Table of Materials). Stir this in a beaker on a magnetic stirring plate at 4 °C until eosin and catalyst are fully dissolved (2 - 3 h).
- Place the samples for 12 - 24 h in the infiltration solution. Keep it at 4 °C under continuous rocking or rotation. Replace infiltration solution with fresh solution after 3 and 12 h. NOTE: Extend infiltration time to 48 h for large embryos (> 1 cm crown/rump length).
- Preparing adult tissues samples (up to 5 x 5 x 7 mm3 )
- Fix the tissues in 2% PFA/PBS containing 4% carbolic acid at 4 °C for at least 3 days. NOTE: Tissue sample stemming from skin, liver, pancreas, kidney, thyroid, heart, striated muscle, brain, nerves and tumor models of several neonatal and adult humans, rodents, pigs, fruit flies, and zebra fishes can be fixed in this solution and then processed for HREM imaging (we demonstrate the fixing of the samples; see Representative Results). Excise the tissue samples with scalpels, scissors or biopsy punches and directly transfer them into fixative. For fixation of nervous tissue use 4% PFA/PBS.
- Remove the fixative and wash the tissue under running tap water for 3-6 h.
- Dehydrate the samples by subsequently placing them in 70%, 80%, 90%, 96% ethanol, mixed with 0.4 g eosin per 100 mL (2 - 3 h each). Keep the solutions with samples at 4 °C under constant rocking or on a rotator. NOTE: For small samples (< 2 x 2 x 2 mm3), the time a sample rests in each ethanol may be reduced to 60 min. For large samples (> 5 x 5 x 5 mm3), it may be extended to 4-8 h.
- Perform all the following infiltration steps under a fume hood wearing protective gloves.
- Prepare the infiltration solution by adding 1.25 g benzoyl peroxide (plasticized catalyst) and 0.4 g eosin to 100 mL of Solution A (for more details, see Table of Materials). Stir in a beaker on a magnetic stirring plate at 4 °C until the eosin and catalyst are fully dissolved (2 - 3 h).
- Place samples in infiltration solution for 24 - 36 h. Keep it at 4 °C under continuous rocking or rotation. Replace infiltration solution with fresh solution after 3 and 12 h. NOTE: For large samples (> 5 x 5 x 5 mm3) extend infiltration time to 48 h.
- Preparing other organic material NOTE: Trim skin substitutes and paper to the desired size with ordinary scissors. No fixation is needed.
- Skip fixation, washing and dehydration steps.
- Perform all the following infiltration steps under a fume hood wearing protective gloves.
- Prepare the infiltration solution by adding 1.25 g benzoyl peroxide (plasticized catalyst) and 0.4 g eosin to 100 mL of Solution A (for more details, see Table of Materials). Stir in a beaker at 4 °C on a magnetic stirring plate until eosin and catalyst are fully dissolved (2 - 3 h).
- Place samples in infiltration solution for 3 - 12 h. Keep it at 4 °C under continuous rocking or rotation. Replace infiltration solution with fresh solution after 1 and 2 - 4 h.
2. Embedding
NOTE: Perform all steps under a fume hood wearing protective gloves.
Prepare fresh infiltration solution as described above (step 1.1.4.1). Add 4 mL Solution B (for details, see Table of Materials) to 100 mL infiltration solution to prepare the embedding solution. NOTE: If precise orientation of the specimen in the embedding molds is required and to allow the embedding of several samples in parallel, slow the polymerization of the stock solution by putting the beaker into an ice-bath.
Use molding cup trays (for details, see Table of Materials) with molds having a routine volume of 6 x 8 x 5 mm3 or 13 x 19 x 5 mm3, or customized molds with volumes of up to 8 x 10 x 15 mm3 (Figure 2).
- Fill the molds with embedding solution and transfer the sample into the mold using a spoon. During transfer ensure that the sample is fully covered with embedding solution to avoid air trapping. NOTE: This can be done at room temperature, if started immediately after collecting the samples and the embedding solution from the refrigerator.
- Orient the sample inside the mold using needles or forceps.
- As soon as embedding solution starts to become viscous, place a block holder (see Table of Materials) on top of the mold and add embedding solution through the central hole of the block holder until the base of the block holder is covered with 1 - 2 mm of embedding solution.
- Seal the molding cup tray by covering it with liquid paraffin wax and wait until it has hardened. Alternatively place a second molding cup tray upside-down on the molding cup tray with samples and seal it with adhesive tape to air-proof.
Allow blocks to finish polymerization by storing the sealed molding cup trays for 1 - 2 days at room temperature.
For post-polymerization processing, place the molding cup trays with the polymerized blocks in a standard laboratory oven and bake at 70 - 80 °C for a minimum of 1 - 2 days until they become dark red. Collect the trays from the oven and remove the blocks from the mold. NOTE: Baking can be done under normal environmental conditions. The covering molding cup tray, which ensures airproof sealing in the first phase after embedding, can be removed to allow for checking the color of the resin after 10 h. Immediately after baking, the resin is soft and the blocks can be easily removed from the molding cup tray. Within 2 - 3 h they are hard and can be stored at room temperature for several months.
3. Data Generation
NOTE: The HREM prototype18,25 used here comprises the following items: (i) Rotary microtome with a block holder that stops after each cut at its upper turning point. (ii) Standard, non-disposable microtome knife, hard metal, profile D (for details, see Table of Materials). (iii) Head of fluorescence compound microscope with objective revolver and a GFP-filter cube (excitation 470/40; emission filter 525/50) in its optical axis. The optical axis is arranged perpendicular to the freshly cut surface of a block mounted on the microtome and is held by a device that allows up- and downward movement. (iv) Motorized cross table carrying the microtome. The table can be shifted in the direction of the optical axis and laterally. (v) Digital video camera attached to the fluorescent compound microscope. (vi) Monochrome light source (470 nm). (vii) Computer connected to the camera, with data generation software (for details, see Table of Materials).
- Procedure
- Indicate the region of interest.
- Use standard laboratory light source (see Table of Materials) for directing white light obliquely to the block surface.
- Identify the outlines of the embedded material as it projects to the block surface. Indicate the region of interest and later field of view on the block surface using a pen or razor blade. NOTE: This step is mandatory to define the extent of the field of view prior to sectioning.
- Define the field of view.
- Mount the resin block with sample on the microtome.
- Move the block holder to its stopping position (upper turning point of block holder excursion).
- Start the digital camera and data generation software, and acquire a live image.
- Choose an objective lens, with an appropriate magnification to cover the region of interest indicated on the block surface. NOTE: Objectives with magnifications 2.5X, 5X, 10X, 20X and numeric apertures between 0.07 and 0.40 are commonly used.
- Move the optic up- and downward and the microtome laterally using the motorized cross table until the region of interest matches with the field of view displayed on the computer screen.
- Focus.
- Perform step by step manual sectioning until first structures of the sample become visible.
- Arrange the block surface in the focus plane of the optic, by moving the microtome in the direction of the optical axis.
- Choose section thickness; section thicknesses from 0.5 to 5 µm are recommended.
- Data generation and processing
- Start software routine that orchestrates sectioning and image capturing. Follow the documented instruction of the imaging capturing software.
- Stop software routine and put a scaling slide in front of the block surface as soon as the sample is completely sectioned.
- Capture an image interactively for later calibration. NOTE: This must be done every time an objective is used in a newly arranged set up.
- Convert the image series to 8 bit gray-scale images in.jpg format.
- Load image series into 3D visualization software and adjust contrasts. NOTE: Image conversion and 3D visualization can be done with various commercially or freely available standard software packages, following the instructions of the respective software.
Representative Results
HREM generates series of inherently aligned digital images with contrasts similar to images of hematoxylin/eosin stained histological sections. As opposed to 2D section images, stacks of HREM images allow for visualization and analysis of the tissue architecture, morphology, and topology of a broad variety of organic materials in 3D. The high contrast often facilitates quick and simple 3D visualization of volume rendered computer models and semiautomatic contour findings for the generation of surface rendered models.
The size of HREM data varies according to the size of the camera target, the mode of image capturing (8, 12, 16 bit, gray scale, color) and the number of single block face images. Smaller data sets of 1,000 8-bit.jpg gray scale images of 2,048 pixels x 2,048 pixels have a size of about 900 MB. Larger data sets of 3,000 8-bit.jpg gray scale images of 4,096 pixel x 4,096 pixel have volumes of approximately 20 GB.
The protocols provided are simple and robust, and during the last decade, they were employed for generating HREM data of many different specimens. Protocol section 1.1 was optimized for processing whole embryos of biomedical model organisms with a length of up to 1 cm and embryo tissue samples with a dimension of up to 5 x 5 x 5 mm3; we used this method for producing HREM data of embryos of the following species: mouse (Mus musculus), chick (Gallus gallus), zebra fish (Danio rerio), quail (Coturnix coturnix), African clawed frog (Xenopus laevis), horse (Equus ferus caballus), milkweed bug (Oncopeltus fasciatus), crocodile (Crocodylia), and octopus (Octopus vulgaris). The data of all species were of excellent quality (e.g., Figure 3)
Protocol section 1.2 was optimized for processing juvenile and adult tissue samples with dimensions up to 5 x 5 x 7 mm3 and employed for visualizing tissue samples of the skin, liver, pancreas, kidney, thyroid, heart, striated muscle, brain, nerves and tumor models harvested from humans (Homo sapiens), mice (Mus musculus), rats (Rattus norvegicus), pigs (Sus scrofa domestica) ferret (Mustela furo), fruit fly (Drosophila melanogaster), and zebra fish (Danio rerio). While the results were excellent with most specimens (e.g., Figure 4, Animation 1), the very central parts of skin (with epidermis) and brain specimens larger than 3 x 3 x 3 mm3 often remained unstained due to insufficient penetration of eosin through these tissues.
Protocol section 1.3 was optimized for processing fibrous organic material and employed for visualizing the fiber architecture of coated paper, uncoated paper, native dermal substitute material and stem cell seeded dermal substitute material. These samples were easy and quick to process. The data of the uncoated paper and most skin substitutes were of good quality (Figure 5, Animation 2). Problems occurred in processing the coated paper, when the anorganic material hindered penetration of eosin. Another issue occurred when processing the skin substitutes on the basis of agar because they became partly digested by the infiltration solution.
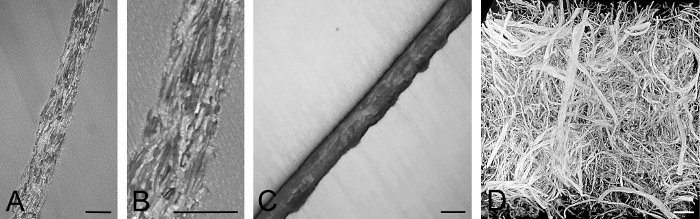
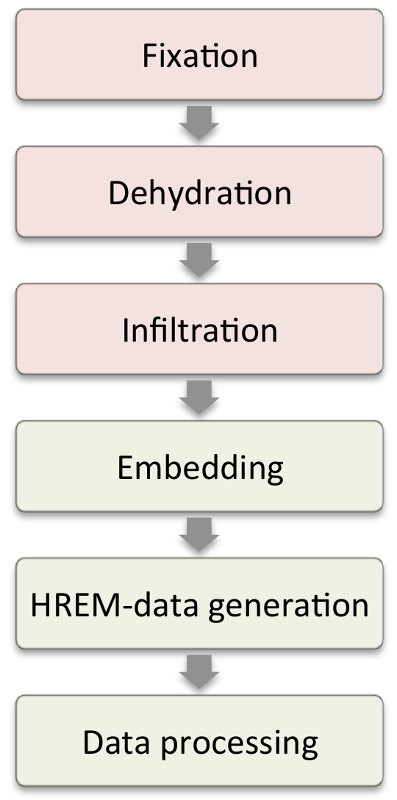
Figure 1: Workflow. Steps shown in the red boxes require modifications according to sample characteristics. Steps in the green boxes are those that are similar in all samples. Please click here to view a larger version of this figure.
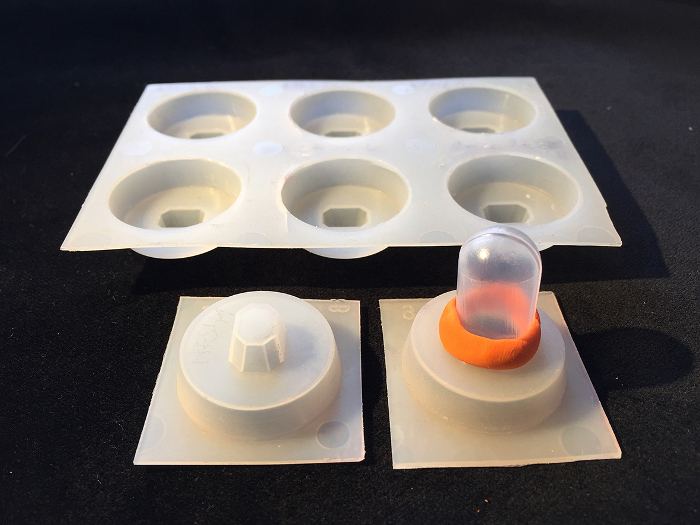
Figure 2: Customized Molds. Molds can be adapted by cutting a hole into the original mold, inserting the bulb of a Pasteur pipet and sealing it with modeling material. Please click here to view a larger version of this figure.
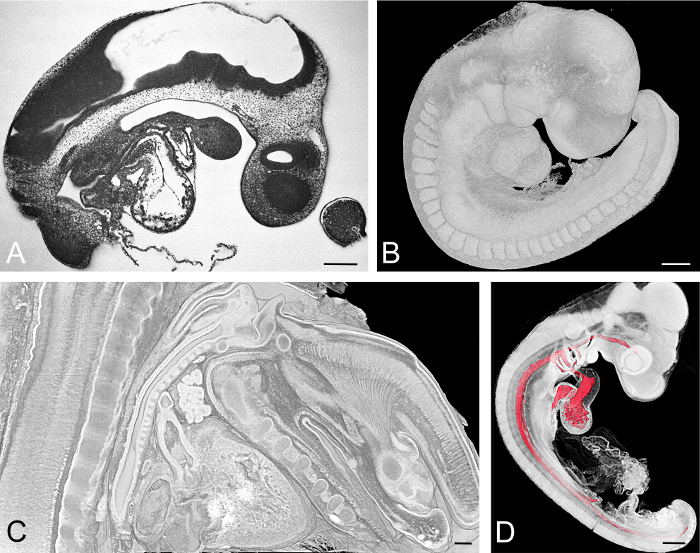
Figure 3: Exemplarily Visualized Embryos.(A, B) Mouse embryo harvested at embryonic day, E=9.5. HREM section image is shown in (A). Volume rendered 3D model showing the surface is shown in (B). (C) Virtual sagittal section through a volume rendered model of the neck of an E15.5 mouse embryo. (D) Chick embryo at developmental Hamburger Hamilton (HH) stage 18. Surface model of the lumina of the cardiovascular components combined with volume rendering of all embryo tissues. Scale bars = 200 µm. Please click here to view a larger version of this figure.
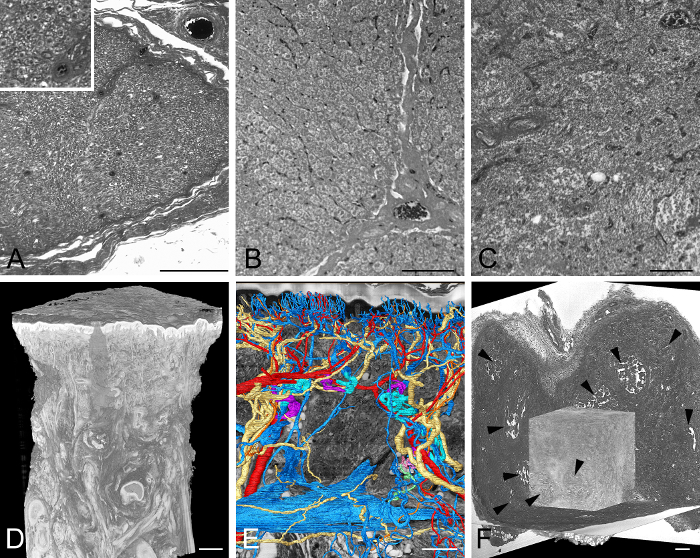
Figure 4: Exemplarily Visualized Adult Tissue Samples. (A) Part of an HREM section image through a human nerve. Inlay shows a section of the image in more detail. (B) Part of an HREM section image through porcine liver. Note the nuclei. (C) Part of an HREM section image of human lymphatic tissue. (D, E) Thick skin of a human thumb pad. Volume rendered 3D model of the entire biopsy. (D) Surface rendered models of arteries, veins, and nerves in front of a virtual resection through HREM data (E). (F) Experimental tumor in adult mouse tissue. Volume rendered 3D model in front of three virtual sections through HREM data. Note the necrotic parts (arrowheads). Scale bars = 200 µm. Please click here to view a larger version of this figure.
Figure 5: Exemplarily Visualized Fibrous Material.(A, B) HREM section image of uncoated, brown paper. (B) shows a section of (A). Note the fibers and their lumen. (C) HREM section of coated paper. Note that the fibers remain unstained. (D) Volume rendered model of native dermal substitute material. Note the different caliber and form of the fibers. Scale bars = 100 µm. Please click here to view a larger version of this figure.
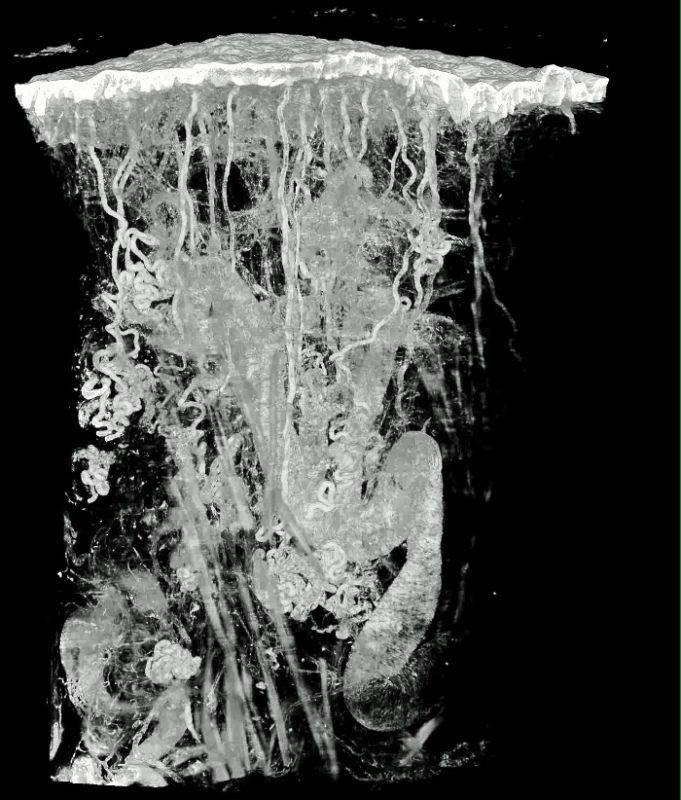 Animation 1: Volume Rendered Model of Thick Skin of a Human Thumb Pad. The size of the tissue block is approximately 4.2 x 2.7 x 2.7 mm3. Voxel size is 1.07 x 1.07 x 2 µm3. Please click here to view this video. (Right-click to download.)
Animation 1: Volume Rendered Model of Thick Skin of a Human Thumb Pad. The size of the tissue block is approximately 4.2 x 2.7 x 2.7 mm3. Voxel size is 1.07 x 1.07 x 2 µm3. Please click here to view this video. (Right-click to download.)
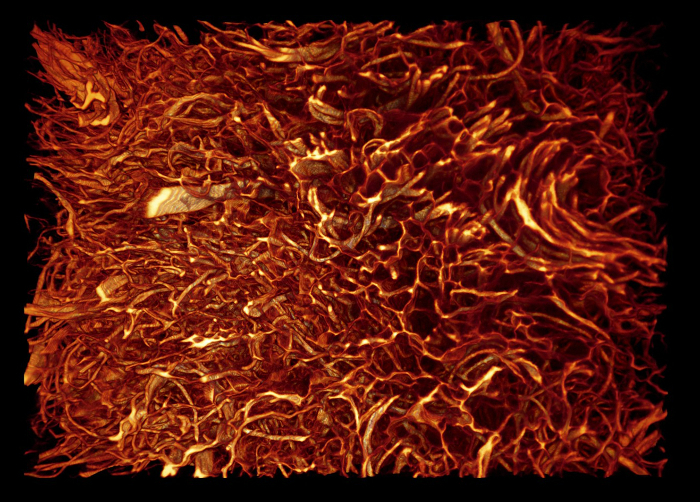 Animation 2: Volume Rendered Model of Native Dermal Substitute Material. The size of the sample is approximately 1.2 x 0.85 x 0.4 mm3. Voxel size is 0.54 x 0.54 x 2 µm3. Please click here to view this video. (Right-click to download.)
Animation 2: Volume Rendered Model of Native Dermal Substitute Material. The size of the sample is approximately 1.2 x 0.85 x 0.4 mm3. Voxel size is 0.54 x 0.54 x 2 µm3. Please click here to view this video. (Right-click to download.)
Discussion
HREM is a highly robust microscopic method that is ideal for visualizing a broad spectrum of organic materials used in biomedicine and industry18,21,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40. It can be employed as an exclusive imaging modality, as currently used by the Mechanisms of Developmental Disorders (DMDD) program41,42,43,44, or as an integrative part of multimodal imaging pipelines45.
A fully functioning HREM data generation apparatus can be assembled from conventional laboratory components and comprises a motorized microtome, microscope, motorized cross table, and a computer with appropriate software25. It is critical to use a microtome equipped with a block holder that reproducibly stops after each section at a defined position and GFP filter cubes inside the optical pathway. However, fully functioning all-inclusive solutions can be purchased from companies such as Indigo Scientific.
HREM faces the same limitations as all histological techniques, except that no artifacts are introduced during sectioning or section mounting. However, there are limitations, which result from the necessity to stain specimens prior to sectioning and from the characteristics of the embedding material. Penetration of eosin through the entire specimen is required for obtaining sufficient tissue contrasts; very dense material, adipose tissues and anorganic substances effectively hinder eosin penetration and this results in unstained tissues in the center of the objects. Using special fixatives helps stain skin specimens, but there is still no proper method for fully overcoming the problem. Another limitation is that resins that block higher than 2 cm tend to break during sectioning. This can be partly avoided by cutting the specimens and processing parts separately.
Correct positioning of small samples or samples with irregular surfaces in the molds during embedding is often problematic. Covering the samples with agarose and processing the agarose blocks as described in the protocol usually resolves this issue19. An alternative approach, which also helps if blocks break during sectioning, is to remove the already hardened block from its holder and embed it anew, following the described embedding procedure.
A typical HREM data set comprises 500 to 3,000 single images. Its numerical resolution is determined by the distance between the successive images (i.e., by section thickness), the characteristic of the camera target, and the properties of the utilized optics. We used section thicknesses between 1 µm and 5 µm and achieved good results, although the presented protocols do not entirely eliminate shining from artifacts20,46. These artifacts are caused by intensely stained tissues located deep inside the block, resulting in blurring of tissue information on the block surfaces by.
The cameras had target dimensions of 2,560 x 1,920 pixels2, 2,048 x 2,048 pixels2, and 4,096 x 4,096 pixels2 and were combined with 1.25X, 2.5X, 5X, 10X, and 20X objective lenses. This resulted in numeric pixel sizes between 0.18 x 0.18 µm2 and 5.92 x 5.92 µm2, which proved to be sufficient for 3D analysis of tissue architecture and cell shapes, and even for visualizing nuclei. Given the high numeric resolution, other cell organelles should be visible as well. Insufficient contrasts due to simple eosin staining, and the optical properties of the objectives dramatically lower the possibility to discriminate structures. The maximal true spatial resolution of the HREM data, which takes into account the numeric aperture, is approximately 1 x 1 x 1 µm3, and therefore only permits effective discrimination of structures larger than approximately 3 x 3 x 3 µm3.
A common problem to all digital imaging techniques is the tradeoff between the size of the field of view, which defines the part of the specimen that can be displayed on the camera target, and the numeric resolution of the image. The larger the field of view, the lower the maximal possible numeric resolution46. The HREM setup used here permits the generation of HREM data with a field of view between 0.74 x 0.74 mm2 (20X objective) displayed in a numeric resolution of 0.18 x 0.18 µm2 and 12.12 x 12.12 mm2 (1.25X objective) displayed in a numeric resolution of 2.96 x 2.96 µm2. Alternative, commercialized set-ups can provide larger fields of views, but at the cost of true resolution. Nevertheless, they provide excellent results, as obvious from the data displayed on the homepage of the DMDD program47.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank Tim Mohun for his invalubale contributions in developing HREM and Petra Heffeter for providing samples.
References
- Murray KK, Seneviratne CA, Ghorai S. High resolution laser mass spectrometry bioimaging. Methods. 2016;104:118–126. doi: 10.1016/j.ymeth.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzlechner M, et al. In Situ Characterization of Tissue-Resident Immune Cells by MALDI Mass Spectrometry Imaging. J Proteome Res. 2017;16(1):65–76. doi: 10.1021/acs.jproteome.6b00610. [DOI] [PubMed] [Google Scholar]
- Elsayad K, et al. Spectrally coded optical nanosectioning (SpecON) with biocompatible metal-dielectric-coated substrates. Proc Natl Acad Sci U S A. 2013;110(50):20069–20074. doi: 10.1073/pnas.1307222110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris FC, et al. A coming of age: advanced imaging technologies for characterising the developing mouse. Trends Genet. 2013;29(12):700–711. doi: 10.1016/j.tig.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Schneider JE, et al. High-resolution imaging of normal anatomy, and neural and adrenal malformations in mouse embryos using magnetic resonance microscopy. J Anat. 2003;202(2):239–247. doi: 10.1046/j.1469-7580.2003.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J. Optical projection tomography as a new tool for studying embryo anatomy. J Anat. 2003;202(2):175–181. doi: 10.1046/j.1469-7580.2003.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD. MicroCT for developmental biology: A versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn. 2009;238(3):632–640. doi: 10.1002/dvdy.21857. [DOI] [PubMed] [Google Scholar]
- Syed SH, Larin KV, Dickinson ME, Larina IV. Optical coherence tomography for high-resolution imaging of mouse development in utero. J Biomed Opt. 2011;16(4):046004. doi: 10.1117/1.3560300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EZ, et al. Multimodal photoacoustic and optical coherence tomography scanner using an all optical detection scheme for 3D morphological skin imaging. Biomed Opt Express. 2011;2(8):2202–2215. doi: 10.1364/BOE.2.002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, et al. Applicability, usability, and limitations of murine embryonic imaging with optical coherence tomography and optical projection tomography. Biomed Opt Express. 2016;7(6):2295–2310. doi: 10.1364/BOE.7.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger WJ, Meng S, Streicher J, Müller GB. A new episcopic method for rapid 3-D reconstruction: applications in anatomy and embryology. Anat Embryol (Berl) 1998;197(5):341–348. doi: 10.1007/s004290050144. [DOI] [PubMed] [Google Scholar]
- Weninger WJ, Mohun T. Phenotyping transgenic embryos: a rapid 3-D screening method based on episcopic fluorescence image capturing. Nat Genet. 2002;30(1):59–65. doi: 10.1038/ng785. [DOI] [PubMed] [Google Scholar]
- Khairy K, Keller PJ. Reconstructing embryonic development. Genesis. 2011;49(7):488–513. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- Medalia O, et al. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298(5596):1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Berrios-Otero CA, Wadghiri YZ, Nieman BJ, Joyner AL, Turnbull DH. Three-dimensional micro-MRI analysis of cerebral artery development in mouse embryos. Magn Reson Med. 2009;62(6):1431–1439. doi: 10.1002/mrm.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, et al. Dual modality optical coherence and whole-body photoacoustic tomography imaging of chick embryos in multiple development stages. Biomed Opt Express. 2014;5(9):3150–3159. doi: 10.1364/BOE.5.003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The morphology of heart development in Xenopus laevis. Dev Biol. 2000;218(1):74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- Weninger WJ, et al. High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol. 2006;211(3):213–221. doi: 10.1007/s00429-005-0073-x. [DOI] [PubMed] [Google Scholar]
- Mohun TJ, Weninger WJ. Embedding Embryos for High-Resolution Episcopic Microscopy (HREM) Cold Spring Harb Protoc. 2012;2012(6):678–680. doi: 10.1101/pdb.prot069583. [DOI] [PubMed] [Google Scholar]
- Geyer SH, Mohun TJ, Weninger WJ. Visualizing vertebrate embryos with episcopic 3D imaging techniques. ScientificWorldJournal. 2009;9:1423–1437. doi: 10.1100/tsw.2009.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneel B, et al. Imaging the zebrafish dentition: from traditional approaches to emerging technologies. Zebrafish. 2015;12(1):1–10. doi: 10.1089/zeb.2014.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, et al. Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening. Dis Model Mech. 2013;6(3):571–579. doi: 10.1242/dmm.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Chew TL. Light sheet microscopes: Novel imaging toolbox for visualizing life's processes. Int J Biochem Cell Biol. 2016;80:119–123. doi: 10.1016/j.biocel.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Powell KA, Wilson D. 3-dimensional imaging modalities for phenotyping genetically engineered mice. Vet Pathol. 2012;49(1):106–115. doi: 10.1177/0300985811429814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer SH, Mohun TJ, Kamolz LP, Weninger WJ. High resolution episcopic microscopy - current applications. Current Biotechnology. 2012;1(4):281–286. [Google Scholar]
- Weninger WJ, Maurer B, Zendron B, Dorfmeister K, Geyer SH. Measurements of the diameters of the great arteries and semi-lunar valves of chick and mouse embryos. J Microsc. 2009;234(2):173–190. doi: 10.1111/j.1365-2818.2009.03159.x. [DOI] [PubMed] [Google Scholar]
- Geyer SH, Maurer B, Dirnbacher K, Weninger WJ. Dimensions of the great intrathoracic arteries of early mouse fetuses of the C57BL/6J strain. The Open Anatomy Journal. 2012;4(1):1–6. [Google Scholar]
- Geyer SH, Maurer B, Pötz L, Singh J, Weninger WJ. High-Resolution Episcopic Microscopy Data-Based Measurements of the Arteries of Mouse Embryos: Evaluation of Significance and Reproducibility under Routine Conditions. Cells Tissues Organs. 2012;195(6):524–534. doi: 10.1159/000329501. [DOI] [PubMed] [Google Scholar]
- Geyer SH, Weninger WJ. Some Mice Feature 5th Pharyngeal Arch Arteries and Double-Lumen Aortic Arch Malformations. Cells Tissues Organs. 2012;196(1):90–98. doi: 10.1159/000330789. [DOI] [PubMed] [Google Scholar]
- Kee HJ, et al. Ret finger protein inhibits muscle differentiation by modulating serum response factor and enhancer of polycomb1. Cell Death Differ. 2012;19(1):121–131. doi: 10.1038/cdd.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer SH, Nohammer MM, Tinhofer IE, Weninger WJ. The dermal arteries of the human thumb pad. J Anat. 2013;223(6):603–609. doi: 10.1111/joa.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer SH, Weninger WJ. Metric characterization of the aortic arch of early mouse fetuses and of a fetus featuring a double lumen aortic arch malformation. Ann Anat. 2013;195(2):175–182. doi: 10.1016/j.aanat.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Maurer B, Geyer SH, Weninger WJ. A Chick Embryo With a yet Unclassified Type of Cephalothoracopagus Malformation and a Hypothesis for Explaining its Genesis. Anat Histol Embryol. 2013;42(3):191–200. doi: 10.1111/ahe.12002. [DOI] [PubMed] [Google Scholar]
- Geyer SH, et al. High-Resolution Episcopic Microscopy (HREM): A Tool for Visualizing Skin Biopsies. Microsc Microanal. 2014;20(5):1356–1364. doi: 10.1017/S1431927614013063. [DOI] [PubMed] [Google Scholar]
- Weninger WJ, et al. Phenotyping structural abnormalities in mouse embryos using high-resolution episcopic microscopy. Dis Model Mech. 2014;7(10):1143–1152. doi: 10.1242/dmm.016337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer SH, et al. High-resolution episcopic microscopy (HREM): A useful technique for research in wound care. Ann Anat. 2015;197:3–10. doi: 10.1016/j.aanat.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Captur G, et al. Morphogenesis of myocardial trabeculae in the mouse embryo. J Anat. 2016;229(2):314–325. doi: 10.1111/joa.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R, Geyer S, Weninger W, Guimberteau JC, Wong JK. The dynamic anatomy and patterning of skin. Exp Dermatol. 2016;25(2):92–98. doi: 10.1111/exd.12832. [DOI] [PubMed] [Google Scholar]
- Jenner F, et al. The embryogenesis of the equine femorotibial joint: The equine interzone. Equine Vet J. 2015;47(5):620–622. doi: 10.1111/evj.12321. [DOI] [PubMed] [Google Scholar]
- Wiedner M, et al. Simultaneous dermal matrix and autologous split-thickness skin graft transplantation in a porcine wound model: a three-dimensional histological analysis of revascularization. Wound Repair Regen. 2014;22(6):749–754. doi: 10.1111/wrr.12233. [DOI] [PubMed] [Google Scholar]
- Mohun T, et al. Deciphering the Mechanisms of Developmental Disorders (DMDD): a new programme for phenotyping embryonic lethal mice. Dis Model Mech. 2013;6(3):562–566. doi: 10.1242/dmm.011957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, et al. Highly variable penetrance of abnormal phenotypes in embryonic lethal knockout mice. Wellcome Open Res. 2016;1:1. doi: 10.12688/wellcomeopenres.9899.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, McGuire C, Mohun T. Deciphering the mechanisms of developmental disorders: phenotype analysis of embryos from mutant mouse lines. Nucleic Acids Res. 2016;44(D1):D855–D861. doi: 10.1093/nar/gkv1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieles G, et al. microMRI-HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos. J Anat. 2007;211(1):132–137. doi: 10.1111/j.1469-7580.2007.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger WJ, Geyer SH. Episcopic 3D Imaging Methods: Tools for Researching Gene Function. Curr Genomics. 2008;9:282–289. doi: 10.2174/138920208784533601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering the Mechanisms of Developmental Disorders: DMDD. 2017. Available from: https://dmdd.org.uk/ [DOI] [PMC free article] [PubMed]


