Abstract
Nucleotide-binding proteins, such as protein kinases, ATPases and GTP-binding proteins, are among the most important families of proteins that are involved in a number of pivotal cellular processes. However, global study of the structure, function, and expression level of nucleotide-binding proteins as well as protein–nucleotide interactions can hardly be achieved with the use of conventional approaches owing to enormous diversity of the nucleotide-binding protein family. Recent advances in mass spectrometry (MS) instrumentation, coupled with a variety of nucleotide-binding protein enrichment methods, rendered MS-based proteomics a powerful tool for the comprehensive characterizations of the nucleotide-binding proteome, especially the kinome. Here, we review the recent developments in the use of mass spectrometry, together with general and widely used affinity enrichment approaches, for the proteome-wide capture, identification and quantification of nucleotide-binding proteins, including protein kinases, ATPases, GTPases, and other nucleotide-binding proteins. The working principles, advantages, and limitations of each enrichment platform in identifying nucleotide-binding proteins as well as profiling protein–nucleotide interactions are summarized. The perspectives in developing novel MS-based nucleotide-binding protein detection platform are also discussed.
Keywords: protein kinases, kinome, affinity purification, protein quantification, multiple-reaction monitoring
I. BACKGROUND
Nucleotide-binding proteins, such as ATP- and GTP-binding proteins, are among the most important families of proteins that are involved in a variety of pivotal cellular processes, including cell signaling (Lemmon & Schlessinger, 2010), proliferation (van Corven et al., 1989), differentiation (Yamamizu et al., 2012), and apoptosis (Reddy et al., 2003). For example, kinases, which represent one of the largest superfamilies of enzymes in higher eukaryotes, mediate cellular protein and lipid phosphorylation to regulate downstream signaling cascade (Manning et al., 2002). Moreover, GTP-binding proteins are also important signal transducing molecules in cells (Bourne, Sanders, & McCormick, 1991). Heterotrimeric G proteins (Preininger & Hamm, 2004) and small GTPases (Takai, Sasaki, & Matozaki, 2001), along with G protein-coupled receptors and protein kinases, mediate important signal-transduction cascades in almost every organism. GTP-binding proteins also encompass various elongation factors, which function in the initiation, elongation, and termination of protein biosynthesis (Bourne, Sanders, & McCormick, 1991).
Despite the importance of nucleotide-binding proteins in cellular functions, the current understanding of nucleotide– protein interactions is far from complete. Individual nucleotide-binding proteins can be characterized using immunoblot (Horscroft & Roy, 2000); however, global study of the structure, function, and expression level of nucleotide-binding proteins in vivo can be hardly achieved with the use of conventional methods owing to the enormous diversity of the nucleotide-binding protein family. Thus, comprehensive identification and quantification of ATP/GTP-binding proteins as well as dynamic analysis of nucleotide–protein interactions at the entire proteome scale are important for understanding better the regulatory mechanisms of nucleotide-binding proteins.
Recent developments in mass spectrometry (MS) instrumentation (Olsen et al., 2009), sample preparation methods (Wisniewski et al., 2009) and bioinformatic tools (Cox & Mann, 2008) have rendered MS-based proteomics a powerful tool for tackling biological and biomedical problems (Bantscheff et al., 2007b). MS has become increasingly the method of choice for analyzing complex protein (Yates, Ruse, & Nakorchevsky, 2009) or metabolite samples (Lorenzo Tejedor et al., 2011; Tsuyama et al., 2008) due to its high specificity, accuracy and throughput. Up to several thousand proteins can be routinely identified and quantified using MS (Choudhary et al., 2009), which could greatly facilitate the extensive study of the perturbations of protein expression in an organism in response to external stimuli (Guo, Xiao, & Wang, 2014). However, proteomic studies of specific family of proteins, including nucleotide-binding proteins, by MS are still a big challenge due to the extreme complexity of the proteome and the relatively low abundance of some proteins. For example, although kinases represent the largest enzyme family in mammals, genes encoding kinases only constitute 1.7% of the human genome (Manning et al., 2002). This limitation can be partially overcome by combining MS with powerful separation techniques, such as polyacrylamide gel electrophoresis (PAGE) (Dong et al., 2011) or multi-dimensional liquid chromatography (Taylor et al., 2009; Wang et al., 2011). Nevertheless, none of these approaches permit selective enrichment of nucleotide-binding proteins from cellular extracts.
In this review, we summarize the recent advances in the use of mass spectrometry, together with affinity enrichment, for the proteome-wide capture, identification and quantification of nucleotide-binding proteins, as well as the global profiling of nucleotide–protein interactions. We also provide perspectives about the future directions in this area of research.
II. GLOBAL KINOME ENRICHMENT/DETECTION PLATFORMS AND THEIR APPLICATIONS
Protein phosphorylation, one of the most important types of post-translational modifications (PTMs), is catalyzed by protein kinases (collectively referred to as the kinome), which are encoded by over 500 genes in higher eukaryotes (Manning et al., 2002). Aberrant expression and/or activation/deactivation of kinases have been implicated as among the main mechanisms through which cancer cells escape normal physiological constraints of cell growth and survival (Blume-Jensen & Hunter, 2001). Additionally, resistance toward cancer chemotherapy is often accompanied with dynamic kinome reprogramming (Duncan et al., 2012). Thus, kinases, because of their crucial roles in cancer development, have become one of the most extensively pursued superfamilies of enzymes as drug targets for cancer chemotherapy, and more than 130 distinct kinase inhibitors have been developed for various phases of clinical trials (Zhang, Yang, & Gray, 2009). Recently, inhibitor potency and selectivity for more than 400 kinases have been reported, which provided a comprehensive target-inhibition profile for the majority of the human kinome (Fabian et al., 2005; Karaman et al., 2008; Davis et al., 2011). Therefore, the kinome-inhibitor interaction networks, coupled with comprehensive profiling of global kinome expression and activity associated with certain types of cancer, could be invaluable for understanding the mechanisms of carcinogenesis and for designing rationally novel kinase-based anti-cancer chemotherapies.
Unfortunately, until recently there had been no optimal method for profiling the expression of the entire kinome at the protein level. Conventional methods for measuring kinase expression rely primarily on antibody-based immunoassays due to their high specificity and sensitivity (Pulford et al., 1997). The immunoassays, however, are restricted by the availability of high-quality antibodies; therefore, these methods are only useful for low-throughput assessment of a small number of kinases. Recent developments in MS instrumentation and bioinformatic tools enabled the identification and quantification of a significant portion of the human proteome from complex samples (Cox & Mann, 2011). However, proteomic studies of the global kinome by MS are still very challenging, which is largely attributed to the fact that, similar as other regulatory enzymes, protein kinases are generally expressed at low levels in cells (Oppermann et al., 2009). This analytical challenge is further exacerbated in shotgun proteomics approach where even more complex mixtures of peptides rather than proteins from cell or tissue extracts are analyzed (Marx, 2013). Therefore, selective enrichment of protein kinases from cellular extracts is essential for the comprehensive identification and quantification of the global kinome.
A. Kinome Enrichment by Affinity Capture Chromatography
Affinity chromatography has emerged as the method of choice for purification of a specific protein or a group of sub-proteome from complex mixtures (Dunham, Mullin, & Gingras, 2012). Although there are more than 500 protein kinases spreading over nine categories, kinases share several common structural and functional attributes (Manning et al., 2002). A very important feature of protein kinases is that most of them employ ATP as the phosphate donor (Shugar, 1996). Therefore, most protein kinases exhibit strong binding toward ATP. Additionally, the available crystal structures of many kinase-ATP complexes showed that most kinases share very similar ATP-binding domains, where the adenine ring of ATP is buried deeply in a hydrophobic cleft, and the α, β, and γ phosphates interact with residues near the mouth of the cleft (Bartlett et al., 2005). The interaction of the adenine ring and α/β phosphates with conserved amino acid residues in kinases is important for ATP binding (Bartlett et al., 2005), hence an ATP analog bearing a modified γ phosphate mimics the native nucleoside triphosphate for investigating the interactions between ATP and kinases.
Based on these observations, a variety of microbeads coupled with ATP moiety on the surface through the γ-phosphate linkage have been developed by multiple vendors for the purification of protein kinases. In an earlier study, researchers used the phosphate-linked ATP-beads to isolate the entire purine-binding proteome from lysates of mouse and human cells (Graves et al., 2002). In that study, the binding and washing buffers contained low concentrations of nucleotides (1mM AMP, 1mM ADP, and 1mM NADH) to reduce nonspecific binding of non-ATP-binding proteins to the resin. Finally, the ATP-binding proteins on the resin were eluted by using a high concentration of ATP. While elution using 20 mM NADH, AMP, or ADP yielded only low quantities of a small number of proteins, elution with 20 mM ATP yielded an abundant and complex protein sample. Importantly, if the ATP was linked to sepharose through the N6 position of adenine (N6-linked resin), very few proteins could be recovered from protein extracts. This finding is consistent with the indispensable role of an intact adenine ring in interaction with ATP-binding proteins. The use of the ATP-sepharose beads led to the identification of 72 and 15 ATP-binding proteins from mouse and human red blood cells, respectively, which included protein and non-protein kinases, dehydrogenases, DNA ligases, and ATPases (Graves et al., 2002). The same approach was also employed for studying the ATP-binding proteome of plant mitochondria (Ito, Heazlewood, & Millar, 2006).
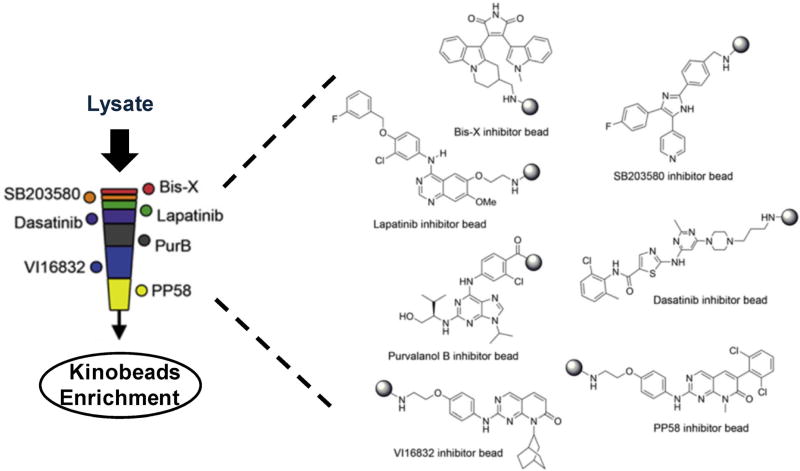
Due to the competitive binding by other ATP-binding proteins, an enrichment method using immobilized ATP as the ligand resulted in the capture of fewer than 10 kinases, and the enriched sample was dominated with highly abundant ATPases, such as heat shock proteins (Graves et al., 2002). To overcome these limitations, Bantscheff et al. (2007a) developed an affinity resin immobilized with a combination of multiple kinase inhibitors, termed kinobeads, to efficiently discriminate protein kinases from other classes of ATP-binding proteins. After screening more than 100 kinase inhibitors, these authors chose seven kinase inhibitors which possess little selectivity and are capable of interacting with kinases from different kinome groups for immobilization on kinobeads. The approach enabled the efficient capture of a large portion of the expressed kinome. In that study, 25mg of lysate samples from HeLa or K562 cells extracts were loaded with a kinobeads suspension. After thorough washes, the bound proteins were eluted with a buffer containing sodium dodecyl sulfate (SDS) as kinase-enriched fraction and separated by SDS–PAGE prior to tryptic digestion and MS analysis. With this method, Bantscheff et al. (2007a) were able to identify, from single pull-down experiments, 174 and 183 protein kinases from HeLa and K562 cells, respectively. These authors also observed a slightly higher coverage for the tyrosine kinase family relative to serine/threonine kinase family, a finding that is agreement with the fact that most of the immobilized kinase inhibitors were designed to target tyrosine kinases. Although kinobeads also bind some ATP- and purine-binding proteins such as chaperones, helicases, and ATPases, it was estimated that kinases represent nearly 80% of all captured proteins based on the total mass spectrometric signal.
Recently, a refined kinase enrichment workflow was proposed by concatenating multiple kinobead resins with different kinase inhibitors as a multi-stage affinity column (Fig. 1), which represents an advance over previous procedures in terms of protein kinase coverage. For example, Duncan et al. (2012) prepared multi-stage kinobead affinity column bearing kinase inhibitors with moderate and poor selectivities (Fig. 1). Daub et al. (2008) developed a similar kinome enrichment strategy by loading cell lysates onto a series of three affinity columns immobilized with five different non-selective kinase inhibitors. A comparison of the total loaded lysate and the eluted kinase-enriched fractions from kinobead affinity columns with gel staining showed that the kinase-enriched fractions only accounted for about 0.3% of the initially loaded amount and a kinase enrichment factor of 100- to 200-fold was achieved (Daub et al., 2008). As a result, the relative expression of more than 200 protein kinases from human cancer cells was routinely quantified using enrichment strategies with these kinobead affinity columns (Daub et al., 2008).
FIGURE 1.
Protein kinase enrichment by multi-column affinity chromatography immobilized with seven different kinase inhibitors. Modified from Duncan et al. (2012).
A variety of kinobeads immobilized with different types of kinase inhibitors have also been developed by multiple research groups. Zhang et al. (2013) introduced a sepharose-supported kinase capture reagent immobilized with a novel kinase inhibitor bisanilino pyrimidine, CTx-0294885, to enable the MS identification of large proportions of the expressed kinome by mass spectrometry. As a complement to traditional kinobeads which generally include pan-kinase inhibitor as the binding bait, Kuster and coworkers synthesized and characterized several more selective affinity resins targeting specific kinase subgroups that were not well represented by the original version of kinobeads (Pachl et al., 2013). Along this line, new affinity probes targeting Akt and many other members of the AGC kinase family (Pachl et al., 2013), fibroblast growth factor receptors (FGFRs) (Ku et al., 2014b), as well as vascular endothelial growth factors and the corresponding receptors (VEGFRs) (Ku et al., 2014a) were developed. For instance, VEGFRs were captured 10- to >100-fold more efficiently with this newly designed VEGFRs affinity resin than using kinobeads, depending on whether tissue or cell extracts were used (Ku et al., 2014a). The combination of pan-kinase probes such as traditional kinobeads with these types of selective kinase affinity resins, may considerably expand the global kinome coverage by affinity capture strategy.
B. Kinome Enrichment by Reactive Affinity Probes
Activity-based protein profiling (ABPP) is the use of chemically reactive affinity probes to study a specific family of subproteome based on their unique structural or functional similarities (Barglow & Cravatt, 2007; Nomura, Dix, & Cravatt, 2010). Therefore, at the heart of the ABPP technology lie the small molecules that are designed to interact with active-site residues of enzymes resulting in the formation of stable covalent bonds. A typical reactive chemical probe usually consists of at least the following three basic elements: (1) a binding group for binding to the active sites of a given class of enzymes; (2) a reactive functional group capable of conjugating with the protein active site to form a stable covalent bond; (3) a chemical tag or reporter group, like biotin and/or a fluorophore, facilitating the rapid isolation and detection of probe-labeled enzymes (Cravatt, Wright, & Kozarich, 2008).
Recently, this activity-based proteomics involving specific labeling of proteins with functional similarities have emerged as an important technique in targeted detection of protein kinases. In this context, the entire proteome is treated with a chemical affinity probe, which incorporates an enrichment tag to a specific functional group on protein kinase family to facilitate downstream purification. For example, in a pioneering study, 5′-p-fluorosulfonylbenzoyladenosine (FSBA), a reactive ATP analog, was employed as an activity-based probe to target nucleotide-binding proteins in whole cell lysates (Hanoulle et al., 2006). Similar to other chemical reactive probes, FSBA harbors three components: a reactive group (fluorosulfonyl), a binding moiety (adenosine), and a tag (sulfonylbenzoic acid). FSBA modifications occur mainly on tyrosine (66.98%) and lysine (32.53%), rarely on serine (0.48%), but not on histidine or cysteine. When applied to the whole proteome, 185 unique FSBA-modification sites in protein lysate of Jurkat cells were identified, including six kinases. Although FSBA is a poor nucleotide mimic displaying weak binding affinity toward ATP-binding sites, this study underscored the potential of activity-based protein profiling in kinome studies.
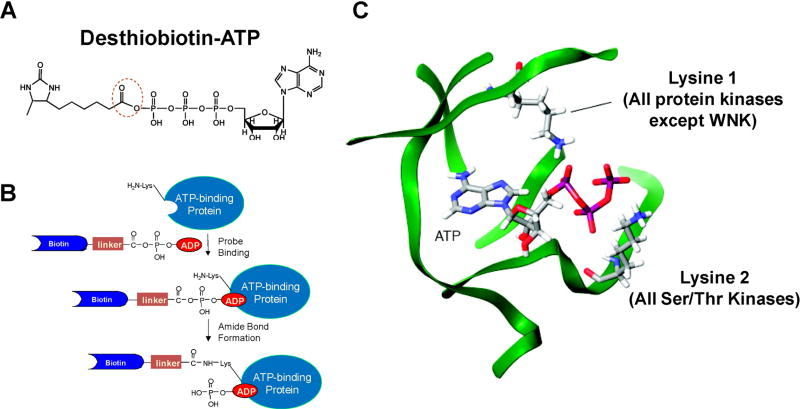
More recently, others and we reported the application of biotin-conjugated acyl nucleotide probe for the enrichment and identification of ATP-binding proteins, especially protein kinases, from complex whole proteome mixtures of different organisms (Fig. 2) (Patricelli et al., 2006; Qiu & Wang, 2007; Ansong et al., 2013; Villamor et al., 2013; Wolfe et al., 2013; Xiao et al., 2013a; Xiao, Guo, & Wang, 2013b). Bioinformatic studies showed that most ATP/GTP-binding proteins carry a consensus amino acid sequence motif termed phosphate-binding loop (P-loop), which constitutes the nucleotide-binding site and confers ATPase/GTPase activity (Saraste, Sibbald, & Wittinghofer, 1990). A conserved motif of GxxxxGK, where “x” represents any amino acid, is often found in the P-loop region of ATP/GTP-binding proteins (Deyrup et al., 1998). In addition, sequence comparisons have shown that virtually all protein kinases have at least one conserved lysine residue within their active sites (Hanks & Hunter, 1995). An invariant lysine residue involved in ATP binding can be localized in a region of subdomain II for all protein kinases except WNK kinases. Furthermore, most kinases, including WNK kinases, possess another conserved motif of HRDxKxxN, which is located in subdomain VIB and participates in ATP binding (Johnson, Noble, & Owen, 1996; Nolen, Taylor, & Ghosh, 2004). Co-crystal structures of protein kinase catalytic domains bound to ATP revealed that these lysine residues are proximal to the β- and γ-phosphates of the bound ATP (Fig. 2C). In light of these previous findings, an ATP analog carrying an acyl phosphate moiety was developed to target the lysine residue in the nucleotide-binding site (Fig. 2A). In the ATP-affinity probe, ATP and biotin (or its analog desthiobiotin) constitute the binding and enrichment moieties, respectively, and they are linked through an acyl phosphate group. The binding of the ATP component of the probe to ATP-binding proteins facilitates the ε-amino group of the P-loop lysine residue to react with the acyl phosphate component to yield a stable amide bond (Fig. 2B).
FIGURE 2.
The principle of reactive ATP affinity probes for kinase enrichment. A: The structure of reactive desthiobiotin-ATP affinity probe. B: A schematic diagram showing the conjugation between reactive ATP affinity probe with an ATP-binding protein. C: The proximity of the two conserved lysine residues to the phosphate groups of ATP. Modified from Patricelli et al. (2006).
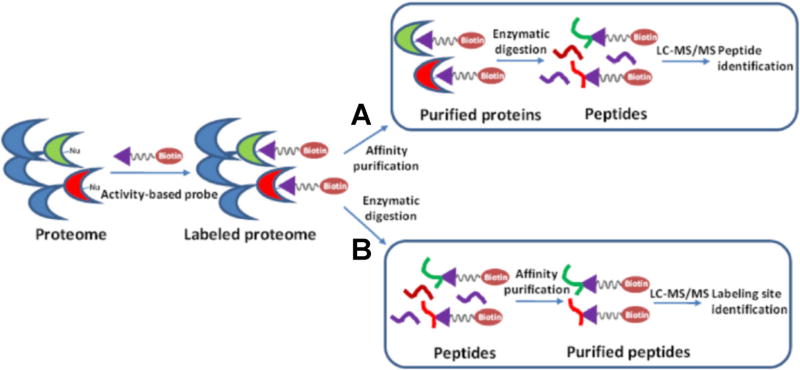
After the labeling reaction, the labeled proteome can be processed in two ways. First, similar as the pull-down procedures employed for kinobeads, the labeled proteins are isolated and enriched by avidin affinity purification. The purified proteins are subsequently digested with trypsin, and the resulting peptide mixtures are analyzed by LC-MS/MS for target identification (Fig. 3). Recently, Lemeer et al. compared the performance of ATP-affinity probe and kinobeads in kinase enrichment, and they found that the two enrichment methods led to the detection of a similar number (~100) of kinases from 1 mg of protein lysate from K562 cells (Lemeer et al., 2013). However, the kinobeads exhibited much better selectivity for kinases than the ATP probe; more than 40% of the total identified mass spectra from kinobeads experiment originate from protein kinases, whereas only 15% of the total identified spectra from the ATP probe experiment are attributed to proteins associated with kinase activity. This difference in selectivity can be simply attributed to the working principles of the two affinity reagents. The ATP probe can potentially label any ATP-binding proteins, whereas the kinobeads are designed to selectively target protein kinases. This notion is further substantiated by the fact that 44% of the identified spectra from the ATP probe experiment belong to proteins with documented nucleotide binding activity, whereas this figure is only 12% in the experiment using kinobeads. In keeping with this finding, the authors also observed that increasing the amount of input material facilitated the detection of more kinases using kinobeads; however, increasing the lysate amount beyond 1 mg in experiments using the ATP/ADP probes led to identification of a large proportion of proteins which were labeled through non-specific interactions (Lemeer et al., 2013). Moreover, a significantly larger number of tyrosine kinases were enriched using kinobeads, whereas members of the STE kinase group (mainly the MAP kinases) were better represented with the use of the ATP probes. This observation is not surprising considering that several of the small-molecule inhibitors immobilized on kinobeads were originally developed to target tyrosine kinases. Based on this observation, an integrated approach encompassing both enrichment methods was developed, where cell lysate was first enriched with the kinobeads and the unbound protein fraction was then subjected to a subsequent pulldown using the ATP probe and avidin agarose. This sequential enrichment led to a 27% increase in the number of unique protein kinases identified (Lemeer et al., 2013).
FIGURE 3.
General procedures for kinase enrichment and detection by activity-based ATP affinity probes. A proteome is first reacted with an ATP-affinity probe, and the labeled proteome can be processed in two ways: (A) The labeled proteins are isolated and enriched by affinity purification. The purified kinases are subsequently digested with trypsin, and the resulting peptide mixtures are analyzed by LC-MS/MS for target identification. (B) The labeled proteins are digested with trypsin to peptides. The biotin-labeled peptides are enriched by affinity purification and analyzed by LC-MS/MS for target identification.
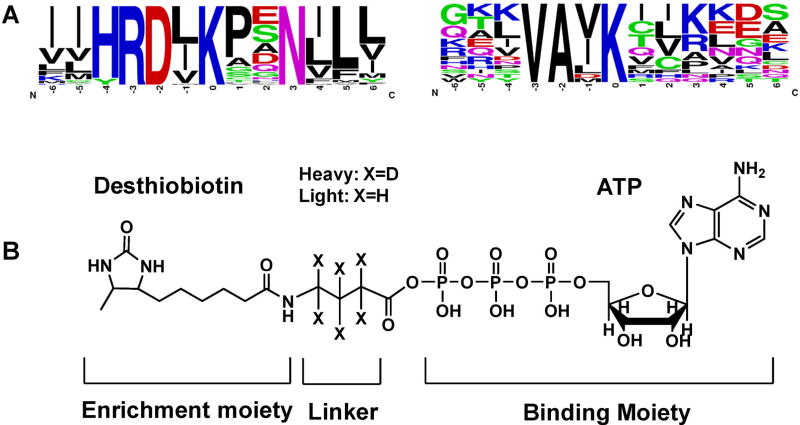
Instead of enrichment at the protein level, the biotin-labeled ATP-binding proteins can be first digested with trypsin to peptides, some of which are probe-modified. The resulting biotin-labeled peptides can then be enriched using avidin agarose and analyzed by LC-MS/MS to determine the identities of the labeled proteins as well as the labeling sites (Fig. 3). In this vein, it is worth noting that biotin conjugation with the side chain of lysine was found not to result in a significant alteration in peptide backbone fragmentation upon low-energy collisional activation or peptide identification based on the resulting MS/MS (Borisov et al., 2002; Sioud et al., 2009). Distinct from kinase capture strategy using kinobeads, this enrichment and analysis workflow for ATP affinity probe-labeled peptides should allow for the determination of lysine residues at the nucleotide binding sites of the enriched proteins, which could potentially provide invaluable information about site-specific interaction between ATP and kinases. For example, two unique sequence motifs of VAxK and HRDxKxxN display a significant enrichment with respect to the occurrence frequency in the desthiobiotin-modified peptides compared to the entire proteome in this high-throughput proteome-wide quantitative analysis (Xiao et al., 2013a; Xiao, Guo, & Wang, 2013b). These two motifs correspond to the two aforementioned invariant lysine residues located at the ATP-binding sites of subdomains II and VIB in kinases, respectively (Fig. 4A). Using this workflow, around 90 protein kinases can be routinely identified in a single proteome (Xiao et al., 2013a; Xiao, Guo, & Wang, 2013b). In addition, in a large-scale analysis of more than 100 human, mouse, rat, and dog proteomes using this workflow with ATP affinity probe, 322 different protein kinases were identified, with 77% (247 kinases) being labeled on one of the two conserved binding-site lysines described above (Patricelli et al., 2006). However, the identification and quantification accuracy of this workflow with enrichment at the peptide level may be somewhat limited viewing that only a single lysine on each kinase is often labeled and thus a single labeled peptide is used for protein determination.
FIGURE 4.
Two unique ATP-binding motifs surrounding biotin-modified lysine residue were found from ATP-probe labeled kinase peptides (A) and the structure of isotope-coded ATP affinity probe (ICAP, B).
Another advantage of enrichment of biotin-labeled kinase peptides lies in its potential application in kinome quantifications. To perform accurate quantitative proteomics study, isotope-labeling techniques, such as stable isotope labeling by amino acids in cell culture (SILAC) (Ong et al., 2002; Ong & Mann, 2006) or isobaric tag for relative and absolute quantitation (iTRAQ) (Ross et al., 2004), are generally required for distinguishing proteins or peptides in different experimental groups. However, the SILAC labeling approach may not be compatible with some applications for clinical samples, for example, biological fluids and tissue samples (Ong et al., 2002), whereas chemical isotopic labeling methods like iTRAQ or dimethyl labeling (Boersema et al., 2009) are prone to additional experimental errors because the isotopic tags are usually incorporated in late stages of the sample preparation. To overcome this limitation, we recently introduced the desthiobiotin-based Isotope-Coded ATP-affinity Probe (ICAP) as an acylating agent to simultaneously enrich and isotopically label protein kinases from the human proteome (Fig. 4B) (Xiao et al., 2013b). The ICAP reagent differs from the aforementioned ATP acyl-phosphate probe in the incorporation of an isotope-coded linker, which is present in light (contains six hydrogens) or heavy (contains six deuterons) form. As a result, the reaction between the probe and the lysine residue at the binding site results in the covalent attachment of desthiobiotin together with light or heavy isotope-coded linker to the lysine residue, which facilitates downstream quantifications. With the use of the ICAP reagents, we successfully quantified the differential expression of 124 protein kinases between human lung tumor and adjacent normal lung tissues from the same patient, clearly demonstrating the potential of the ICAP reagents in quantitative kinome study of clinical samples (Xiao et al., 2014a).
C. Comprehensive Kinome Expression or Activity Profiling Strategies
Kinobeads and reactive ATP affinity probes are considered the two most promising strategies for kinome enrichment, and they exhibit considerable potential for the comprehensive profiling of global kinase expression in response to distinct extracellular stimuli, and for the unambiguous identification of targets of kinase inhibitors by quantitative mass spectrometry.
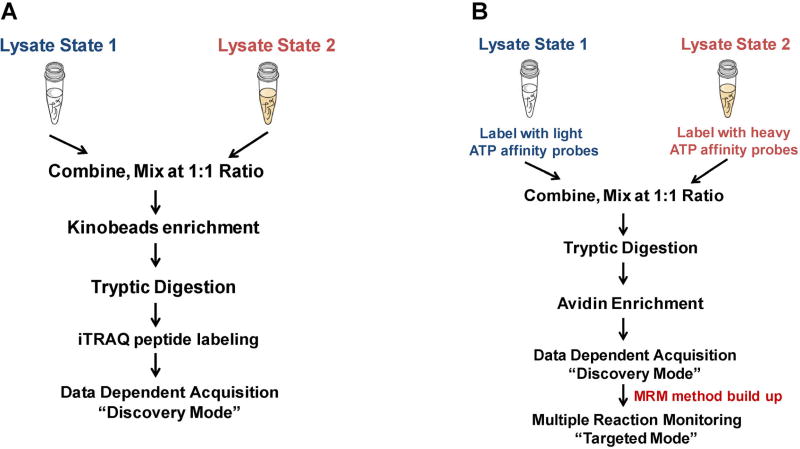
First, the combination of selective kinase enrichment with quantitative MS techniques, such as SILAC (Daub et al., 2008; Oppermann et al., 2009) or iTRAQ (Duncan et al., 2012) could enable comprehensive profiling of the expression and activity of the global kinome. For example, activation of alternative routes of kinase pathway following targeted inhibition of specific kinases is often accompanied with the resistance of kinase-targeted anti-cancer drugs (Johannessen et al., 2010; Chandarlapaty et al., 2011). To explore the dynamic reprogramming of kinome following MEK kinase inhibition, Duncan et al. (2012) assessed the activity and drug responses of a significant percentage (50–60%) of the expressed kinome in triple-negative breast cancer (TNBC) cells and genetically engineered mice using kinobeads. In this study, cells or tumor tissues were lysed on ice with non-denaturing detergents to preserve the binding affinity for interested kinases. Approximately 20–40 mg of protein lysate was loaded to affinity column of layered kinobeads immobilized with seven types of kinase inhibitors. The eluted protein sample was further digested with trypsin and resulting peptides were subjected to iTRAQ labeling to facilitate the quantification. The iTRAQ-labeled peptide mixtures were then analyzed by LC-MS/MS (Fig. 5A).
FIGURE 5.
Common workflows for global kinome analyses. A schematic diagram showing the general workflow for protein kinase analysis facilitated by kinobeads enrichment (A) and multiple-reaction monitoring (MRM) analysis of global kinome using ICAP (B).
The results from the analysis of global kinome in TNBC patient cells showed activated RAF-MEK1/2-ERK1/2 signaling, supporting MEK as a target in TNBC. More importantly, the kinome profiling results indicated that pharmacologic MEK inhibition in TNBC cells and tumors of genetically engineered mice resulted in rapid kinome reprogramming. This occurs through the induction of c-Myc degradation and subsequent expression and activation of multiple tyrosine receptor kinases (TRK) that bypassed the initial MEK-ERK inhibition. These global kinome response signatures upon MEK inhibition allowed the researchers to design and test the efficacy of a novel combination therapy including inhibitors for both MEK and TRK in TNBC treatment, which overcomes effectively single agent resistance.
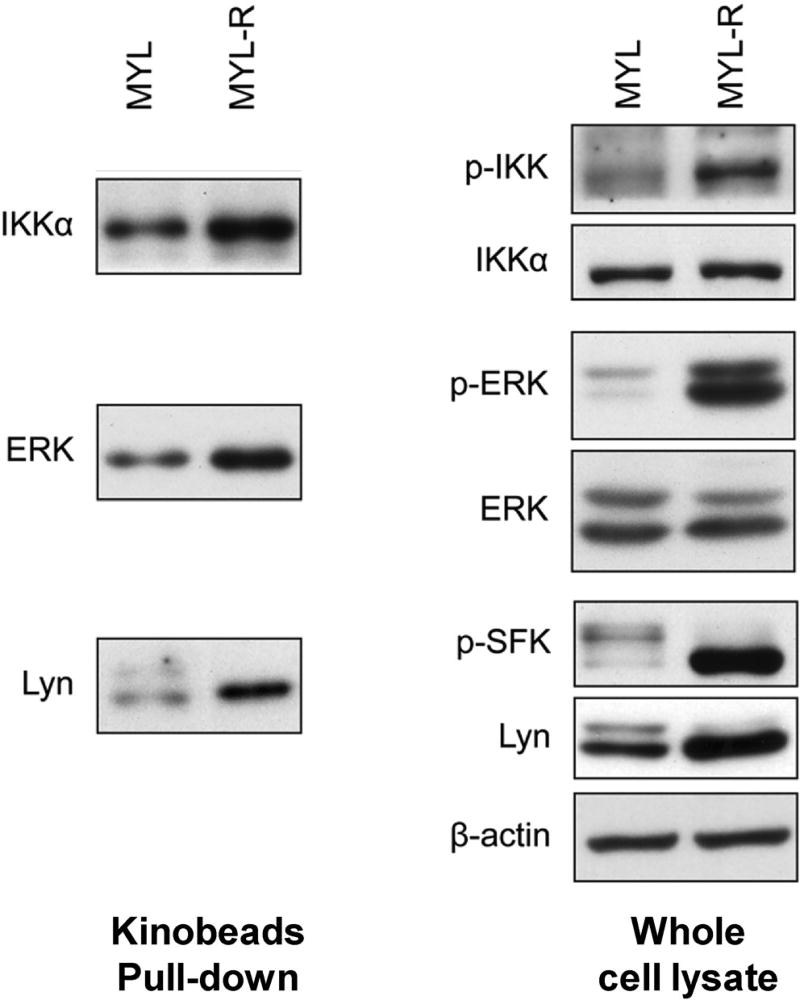
An interesting question in the global kinome enrichment strategy by kinobeads is whether the kinome beads can preferentially capture activated (phosphorylated) over inactivated (unphosphorylated) form of kinases. To investigate the activity-dependent binding of kinobeads, the same group performed pull-down assays to compare the amount of kinases bound to kinobeads with or without phosphatase treatment (Cooper et al., 2013). Treatment of MYL-R lysates with alkaline phosphatase eliminated the binding of IKKα, Lyn and MEK2 to kinobeads, demonstrating the necessity of kinase activity on kinobeads binding. Furthermore, the comparison of the levels of total expression and activated form of kinases, as well as the captured kinases by kinobeads between imatinib resistant- and sensitive-chronic myeloid leukemia cell lines (MYL-R and MYL), revealed that the level of captured kinases by kinobeads was affected by both the expression level and the activation state of the kinase. For example, IKKα, Lyn and ERK displayed elevated capture by kinobeads from the lysates of MYL-R than MYL cells (Fig. 6). Immunoblotting analysis of the whole cell lysate with the use of high-quality phospho-specific antibodies showed that, while the amount of Lyn kinase captured by kinobeads between these two cell lines is correlated with its total expression level, the differences in IKKα and ERK after kinobeads pull-down were solely induced by the alteration in their activation state (Cooper et al., 2013). Therefore, these results suggested that kinobeads can be applied as a powerful tool to profile the changes in the expression and activation of the kinome.
FIGURE 6.
Validation of kinome profiles of MYL and MYL-R cells by immunoblotting. Left, the relative amounts of Lyn, IKKα, ERK kinases in kinobeads pull-down fractions from MYL and MYL-R cell lysates (left); right, total and activated amount of Lyn, IKKα, ERK kinases in whole cell lysates from MYL and MYL-R cells. Modified from Cooper et al. (2013).
When coupled with phosphopeptide enrichment and detection techniques, the aforementioned kinome enrichment strategies can allow for global in-depth survey for the activity of the kinome. For instance, the combination of kinobeads with TiO2 phosphopeptide enrichment was employed to analyze protein kinase regulation in cell-cycle progression (Daub et al., 2008). In that study, activities of 219 protein kinases with more than 1,000 phosphorylation sites from S and M phase-arrested human cancer cells were comprehensively examined and numerous novel M phase-induced activation of kinases with established mitotic functions were revealed (Daub et al., 2008). Similarly, McAllister et al. (2013) successfully compared the activities of more than 200 kinases across six cell lines representing different clinical subtypes of breast cancer by performing kinome capture with ATP reactive probes followed by phosphopeptide enrichment using TiO2.
Despite these advances, the LC-MS/MS data for such large-scale kinome studies are often acquired in the data-dependent acquisition (DDA) mode, where typically 10–20 most abundant ions found in MS are selected for fragmentation in MS/MS to enable peptide identification (Olsen et al., 2009). Although this discovery-mode (or shotgun) proteomic approach provides the potential to uncover novel protein targets, sample complexity, together with inherent variations in automated peak selection in the DDA mode, results in limited sensitivity and reproducibility for protein quantification. As a result, only partially overlapping sets of proteins can be identified even from substantially similar samples (Marx, 2013). The inadequate sensitivity and lack of reproducibility of these kinome detection strategies hamper their utility in biomarker discovery and clinical studies. Targeted proteomics technique, relying on multiple-reaction monitoring (MRM) on triple quadrupole mass spectrometers, has become increasingly used in quantitative proteomics studies (Picotti & Aebersold, 2012). In the MRM mode, mass filtering of both the precursor and product ions is employed to provide high specificity for the quantification of target proteins. Additionally, this MRM-based targeted MS analysis permits rapid and continuous monitoring of specific ions of interest, which enhances the sensitivity for peptide detection by up to 100-fold relative to MS analysis in DDA-based discovery mode (Lange et al., 2008).
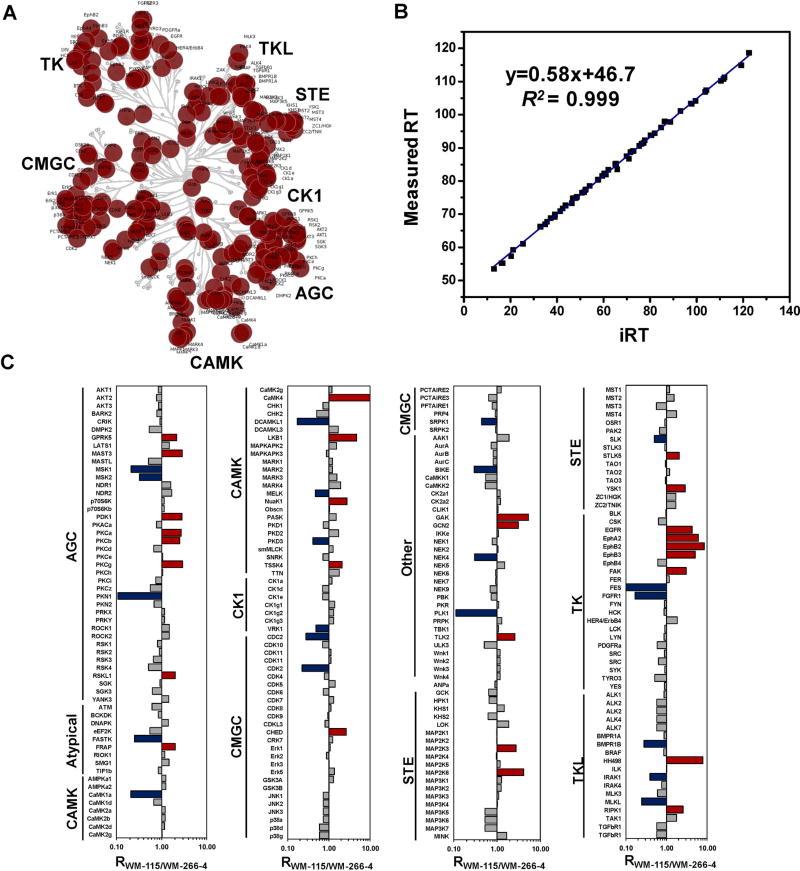
We recently introduced an MRM-based kinome profiling assay, in conjunction with the use of the ICAP probes, for more than 300 human protein and lipid kinases by monitoring specifically targeted peptides located at the ATP-binding sites of kinases (Xiao et al., 2014a). The design of the MRM-based kinome assay requires a priori tandem mass spectral information for the desthiobiotin-modified kinase peptides (Lange et al., 2008). Therefore, we first built up a kinome MRM library based on large-scale discovery-based shotgun MS experiments conducted in our laboratory, which contains a total of 386 peptides from 313 human kinases, including 292 protein kinases, 14 lipid kinases, and 7 metabolic kinases harboring the P-loop GxxxxGK motif. Most targeted kinase peptides in our kinome library contained one of two unique kinase motifs with demonstrated ATP-binding affinity, that is, HRDxKxxN and VAxK, ensuring the high specificity of our MRM-based kinome assay. Placement of these protein kinases to the human kinome map (Manning et al., 2002) revealed that our kinome library spreads over all major groups of the human kinome (Fig. 7A). It is worth noting that, with known motif information, these targeted kinase peptides can be accurately predicted from the amino acid sequences of kinases; therefore, our MRM kinome library can be readily expanded to the whole kinome by generating the retention time signature and tandem mass spectra of synthetic standard peptides for these kinases.
FIGURE 7.
MRM-based targeted proteomic approach for quantitative kinome profiling. A: Targeted protein kinases mapped in the dendrogram of the human kinome. B: iRT values predict measured RT in LC-MRM experiment (130 min linear gradient) with an excellent correlation coefficient (R2 = 0.999). C: Quantitative comparison of kinome expression in WM-115 and WM-266-4 cells. Blue and red bars denote kinase upregulated in WM-266-4 and WM-115 cells, respectively. Modified from Xiao et al. (2014a).
Our current kinome peptide list encompasses 386 peptides. At least three transitions were monitored for each light and heavy labeled peptide, which involves the monitoring of more than 2,000 MRM transitions in quantitative measurements. Successful detection at this level of multiplicity necessitates scheduled MRM analysis in which the mass spectrometer is programmed to monitor a limited number of peptides in predefined retention time windows (Stahl-Zeng et al., 2007). Therefore, our multiplexed MRM-based kinome assays require accurate prediction of retention time (RT) for the kinase peptides. To this end, we calculated the normalized retention time (iRT) for each peptide on our target list following a previously described method (Escher et al., 2012). Based on previous retention time information of targeted peptides on an Orbitrap Velos coupled with EASY-nLC II system, we used 10 BSA peptides as standards to successfully convert empirically determined retention times of 94% (362 out of 386) of targeted peptides into normalized iRT scores, which reflect their conserved elution order (Fig. 7B). Importantly, the incorporation of iRT into MRM kinome library renders our MRM kinome assay easily transferable across different chromatographic systems and laboratories. Facilitated by iRT retention time prediction platform, we can simultaneously monitor approximately 1,000 transitions in a single LC-MS analysis, rendering a complete coverage for the entire kinome library within three LC-MS runs.
With this MRM platform, we demonstrated that ~250 protein kinases (50% of the entire human kinome and more than 80% of human kinome in a single cell line) could be routinely quantified without extensive separation using multi-dimensional chromatography (Xiao et al., 2014a). For example, we applied the MRM-based kinome assay to assess the differential expression of kinases in a pair of human melanoma cell lines, that is, WM-115 and WM-266-4 cells (Xiao et al., 2014a), which were initially derived from the primary and metastatic melanoma sites of the same patient (Westermark et al., 1986). We quantified a total of 246 kinases in WM-115 and WM-266-4 cells (Fig. 7C), including kinases from all seven major kinome groups as well as other and atypical kinases. Moreover, our studies showed that the MRM-based kinome assay displays superior sensitivity, reproducibility and accuracy over discovery-based shotgun proteomics. Along this line, a total of 246 kinases in the library were quantified in MRM-based targeted kinome assay, whereas only 136 protein kinases were quantified by shotgun proteomics strategy even with multi-dimensional LC separation. Moreover, 99% (239 out of 242) of the kinase peptides were successfully quantified in both forward and reverse labeling experiments in MRM-based targeted analysis. By contrast, only 64% (103 out of 161) were commonly quantified in forward and reverse labeling experiments for the same samples using the DDA-based shotgun proteomics approach. Thus, the MRM-based targeted approach afforded much better reproducibility than the DDA-based shotgun approach for kinome profiling analysis in which multiple experimental replicates are generally required. Importantly, our results demonstrated that the anti-proliferative effects of kinase inhibitors are correlated with the expression or ATP-binding affinities of their targeted kinases. In this regard, we found that multiple receptor tyrosine kinases, including EphA2, EphB3, EphB4, were upregulated in WM-115 cells. This is correlated with markedly elevated sensitivity of WM-115 cells toward dasatinib, a potent inhibitor for EphA2, EphB3, and EphB4 (Karaman et al., 2008), than WM-266-4 cells. This finding is reminiscent of a previous study showing that dasatinib could suppress the growth and reduce the migration and invasion of WM-115 cells, but not WM-266-4 cells (Eustace et al., 2011). Therefore, this facile and accurate kinome profiling assay, in combination with the kinome-inhibitor interaction map (Fabian et al., 2005; Karaman et al., 2008; Davis et al., 2011), may provide invaluable knowledge to predict the effectiveness of kinase inhibitor drugs and offer the opportunity for individualized cancer therapy.
D. Determination of the Binding Constants of Kinase Inhibitors Toward Native Kinases
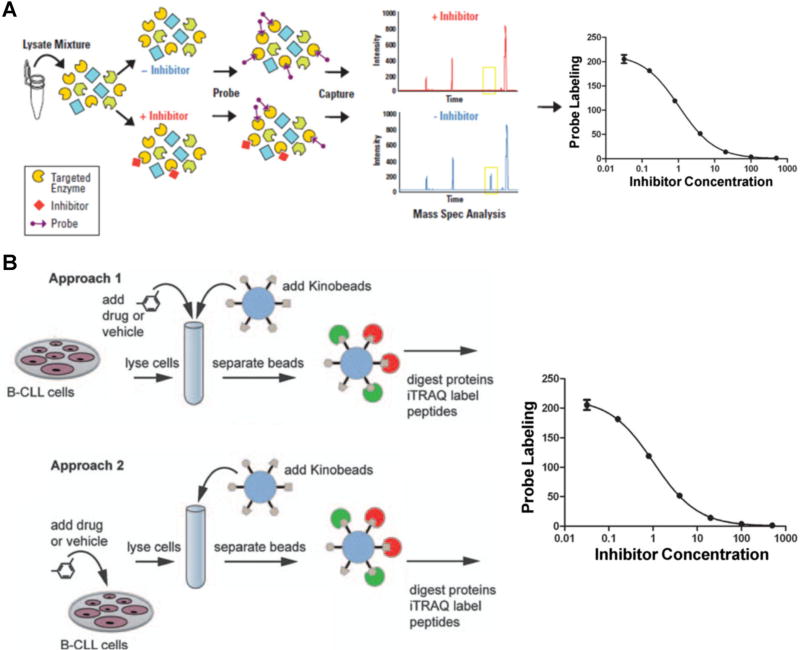
The above-described kinome enrichment strategies also allow for the quantitative determination of the affinity and selectivity profiles for kinase inhibitors in any cell types or primary tissues. Recently, inhibitor potency and selectivity for more than 400 kinases have been reported (Fabian et al., 2005; Karaman et al., 2008; Davis et al., 2011), which provided a comprehensive target-inhibition profile for the majority of the human kinome. However, these data were obtained with the use of the recombinant full-length or catalytically active fragment of kinases. In addition, the biological functions and inhibitor efficacy of many protein kinases were found to depend largely on cellular context. For instance, the activation, deactivation and localization of mitogen-activated protein kinases (MAPKs) are partly determined by scaffolding and anchoring proteins of MAPKs in native cellular environment (Garrington & Johnson, 1999). In addition, post-translational modification (PTM) such as phosphorylation has also been considered as one of the most important factors to regulate the spatial and temporal properties of protein kinases (Newton, 2001). Therefore, kinase-inhibitor assay solely based on recombinant proteins may not recapitulate the complexity of kinase function and regulation in native biological systems and it is highly desirable to perform these protein kinase-inhibitor interaction studies using native protein extracts (Rosenblum, Nomanbhoy, & Kozarich, 2013). To this end, different concentrations of drugs are titrated into a protein extract, and kinase enrichment is subsequently performed using either kinobeads or ATP affinity probes. The free inhibitor in the lysate competes with the affinity matrix binding or affinity labeling for the kinase target; therefore, kinases with high affinity to the free inhibitor display a reduced binding to the affinity matrix or labeling efficiency, which can be identified and quantified by MS. For example, Patricelli et al. (2011) and Rosenblum, Nomanbhoy, and Kozarich (2013) developed an ATP probe-based platform called KiNativ to profile several well-studied kinase inhibitors against >200 kinases in cell lysates. KiNativ analyzed the kinase-enriched samples by a “targeted” MS approach on a linear ion trap mass spectrometer, whereby the instrument is preprogrammed to acquire MS/MS data of known kinase peptide ions in specified retention time windows. Data collected using the kinase target lists were analyzed by extracting characteristic fragment ions for each kinase peptide (Fig. 8A). Similar to the MRM-based analytical platform we developed, they found that the signal/noise ratio of the summed fragment ion traces from the targeted MS/MS spectra were typically 50-fold higher than that of the corresponding parent ion chromatograms in the MS scans. Using inhibitor concentrations at 10, 1, 0.1, and 0.01 µM as a competitor, the apparent Kd value for each inhibitor can be estimated based on the IC50 values measured for the competition between inhibitors and the ATP-affinity probe. Notably, their results revealed that biological targets for some of these inhibitors were strikingly different between native and recombinant kinase inhibitory profiles. For example, several Raf kinase inhibitors exhibited markedly different binding toward native and recombinant Raf-1. The different behaviors observed for these compounds with the use of recombinant proteins and whole cell extracts appear to emanate from differences in the properties and/or conformation of the kinases in a native setting or expressed in isolated recombinant systems.
FIGURE 8.
Kinome profiling assays for the characterizations of the binding affinities and targets of kinase inhibitors. A: Workflow of a KiNativ experiment to determine the binding constants of kinase inhibitors. Whole proteome is treated with an inhibitor followed by addition of the reactive ATP affinity probe. After tryptic digestion and streptavidin enrichment, the probe-labeled kinase peptides with or without inhibitor treatment are quantified by LC-MS analysis. Modified from Patricelli et al. (2011). B: Workflow of kinobeads competition assay to determine drug targets. In the first approach, the drug of interest is added to the lysate at different concentrations followed by enrichment using the kinobeads, where the drug competes with the kinobeads for binding to protein kinases; in the second approach, live cells are kept in culture and treated with various concentrations of drug. Subsequently, cell lysates are generated and endogenous protein kinases are captured by kinobeads and analyzed by LC-MS. Modified from Kruse et al. (2011).
Similar kinase inhibition profiling experiments were also conducted using kinobeads (Bantscheff et al., 2007a; Pachl et al., 2013; Ku et al., 2014a,b). Bantscheff et al. (2007a) assessed quantitatively the binding targets of three ABL tyrosine kinase inhibitors including bosutinib (SKI-606), which is currently used in clinical studies, and the marketed drugs imatinib and dasatinib. In that study, the drugs were added to K562 cell lysates, which express the gene encoding the constitutively active BCR-ABL fusion protein, in concentrations ranging from 100 pM to 10 mM. When the drug in the lysate binds to its target and thus blocks the ATP-binding site, a reduced amount of the free target is available for capture by kinobeads, whereas the binding of non-targeted kinases or other proteins is not affected. The kinobeads-bound material from each spiking experiment was subjected to tryptic digestion and peptides were labeled with different forms of the iTRAQ reagent. Subsequently, peptide mixtures were combined and subjected to MS analysis. Relative protein quantification was achieved by measuring the signal of the iTRAQ reporter ions relative to vehicle-treated lysate. From this data set, the dose-dependent binding profiles could be computed for approximately 150 kinases in each sample.
Recently, a comprehensive strategy was developed to profile and determine the targets of kinase inhibitors by kinobeads (Kruse et al., 2011). The unique feature of this strategy is that both cell lysates and live cells were employed for the assay. As shown in Figure 8B, in the first part of the experiment, the authors conducted the kinobeads competition-binding sassy in combination with iTRAQ labeling and LC-MS/MS to quantitatively assess the binding targets of inhibitor of interest. Similar as described above, kinase inhibitor BMS-387032, at concentrations of 40 nM to 10 µM, was spiked into lysate from patient-derived primary chronic lymphocytic leukemia (CLL) cells. As a consequence, the BMS-387032 binding targets are prevented from binding to the kinobeads in a dose-dependent manner, whereas the binding of other proteins to the kinobeads is not affected. To further validate the candidate targets and discover novel indirect targets, the authors conducted target-profiling experiment using live primary CLL cells in parallel. In this case, BMS-387032 was incubated with living primary CLL cells for 6 h and lysates subjected to kinobeads profiling and LC-MS/MS analysis. As a result, it was found that multiple kinase targets, such as CDK9 and PCTK2, exhibited potent and consistent binding toward BMS-387032 in target profiling assay with the use of cell lysate and living CLL cells.
III. GTP-BINDING PROTEIN ENRICHMENT AND DETECTION PLATFORMS
Guanosine triphosphate-binding proteins are a superfamily of more than 100 proteins that include heterotrimeric G proteins, small GTPases and a variety of other proteins such as elongation factors and tubulins. For instance, the dynamic interplay of GDP-bound inactive and GTP-bound active forms of heterotrimeric G proteins is closely associated with the signal amplitude of G-protein-related pathway (Preininger & Hamm, 2004). The regulation of the activity of small GTPases plays vital roles in cell signaling and their dysregulation is believed to be closely associated with the development of various types of cancer (Cox & Der, 2002). However, the properties and functions of a number of GTP-binding proteins are still unknown because of their relatively low abundances. Hence, functional proteomic analysis of GTP-binding proteins necessitates their efficient enrichment.
Since GTP is a common ligand for all GTPases with reasonable affinity at a conserved P-loop motif sequence (GxxxxGK) (Dever, Glynias, & Merrick, 1987; Saraste, Sibbald, & Wittinghofer, 1990), the obvious choice of a scaffold for GTP affinity beads or probes is GTP. In this connection, GTP-immobilized affinity beads are commercially available, and a variety of reactive GTP analogues as affinity probes have also been developed. For instance, similar as the acyl phosphate-based ATP probe, desthiobiotin-GTP probe was also synthesized to enable selective labeling and enrichment of small GTPases and large G-protein subunits (Xiao et al., 2013a, 2014b). Additionally, Kaneda et al. (2007) developed a simple and efficient photoaffinity method with the use of diazirine-carrying GTP analogue to facilitate proteomic analysis of GTP-binding proteins. Recently, George Cisar and coworkers synthesized and characterized a photoreactive GTP-BP-yne affinity probe, with photoactivatable benzophenone (BP) for cross-linking to GTP-binding protein targets and an alkyne tag for conjugation to reporter tags by using click chemistry. Facilitated by this kind of GTP affinity probe, more than 30 GTP-binding proteins including small GTPases and members of the GTP1/OBG family were identified by MS (George Cisar, Nguyen, & Rosen, 2013).
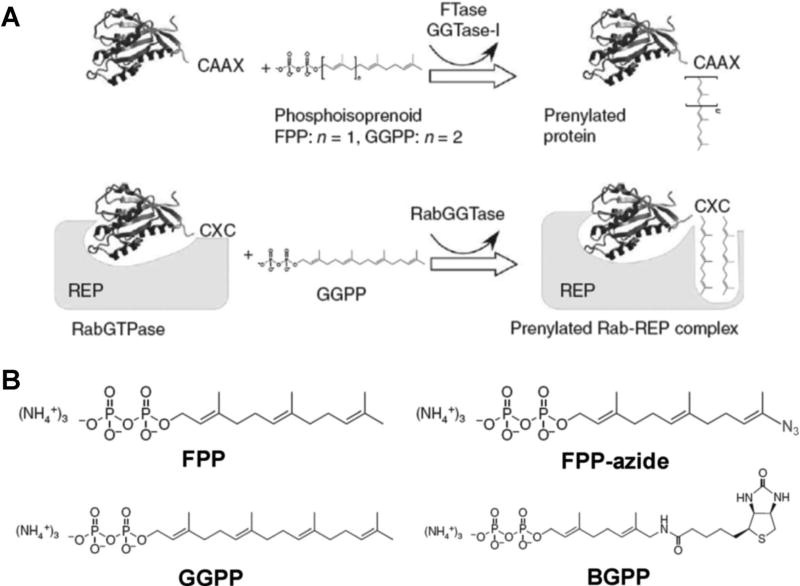
Small GTPases or Ras superfamily, with a molecular weight of ~21 kDa, are among the most important GTP-binding proteins (Cox & Der, 2002). It has been revealed that highly conserved amino acid sequence (30–55% homologous to each other) exists in at least the Ras, Rho, Rab, Sar1/Arf, and Ran families of small GTPases (Takai, Sasaki, & Matozaki, 2001). Aside from the known P-loop sequence motif present in most GTP-binding proteins near the N-termini, there are unique amino acid sequences of CAAX or CXC (“A” and “X” represent aliphatic acid and any amino acid, respectively) near the C-termini of small GTPases. The cysteine residues in the CAAX and CXC motifs may be post-translationally modified with lipids, such as farnesyl or geranylgeranyl functionality through a thioether linkage. These modifications, also known as protein prenylation, facilitate the translocation of Ras protein from the cytosol to cytoplasmic face of the plasma membrane (Casey & Seabra, 1996; Fig. 9A). Based on this unique PTM, an enrichment and detection strategy for small GTPases involving the metabolic incorporation of a synthetic farnesyl or geranylgeranyl analog was developed (Kho et al., 2004). Farnesyl pyrophosphate (FPP) constitutes the farnesyl group donor for post-translational modification of Ras protein. Thus, an FPP analogue, FPP-azide, was synthesized and incubated with tumor cells for metabolic labeling (Fig. 9B). Due to the structural similarity of FPP and FPP-azide, no difference in enzymatic farnesylation of FPP-azide and FPP was observed. Next, the F-azide-modified proteins were selectively ligated to a phosphine biotin probe via the Staudinger reaction. Finally, the farnesy-lated protein-phosphine biotin conjugates were affinity purified using avidin beads and detected by LC-MS. A total of 18 proteins in the Ras superfamily, including H-ras, K-ras, N-ras, Rheb, and Rap2, were identified with this approach. Recently, Nguyen et al. (2009) described another type of functionalized isoprenoid, biotin-geranylpyrophosphate (BGPP) as the protein prenylation substrate analogue. BGPP can be recognized by wild-type Rab geranylgeranyltransferase (RabGGTase) as well as engineered protein farnesyltransferase (FTase) or geranylger-anyltransferase-I (GGTase-I). Therefore, all prenylatable proteins can be modified by BGPP, allowing for rapid isolation and characterization of the entire “prenylome,” which include a large percentage of Ras superfamily (Fig. 9B). Using this approach, they successfully quantified the relative abundances of almost all the 42 members of the Rab GTPase family in COS-7 cells (Nguyen et al., 2009). More importantly, this approach allows for the characterization of the effects of protein prenyltransferase inhibitors such as BMS3 on these RabGT-Pases in vivo, which differ from previous observations made in in vitro experiments (Nguyen et al., 2009).
FIGURE 9.
Chemical proteomic method for the characterizations of protein prenylation. A: A scheme of protein prenylation by formation of a thioether linkage between the prenyl group and one or two C-terminal cysteines of the protein substrate. Prenylation reaction can be catalyzed by the CAAX prenyltransferases FTase and GGTase-I or CXC prenyltransferase RabGGTase. B: The chemical structure of FPP-azide and BGPP in comparison with the natural prenylation substrates FPP and GGPP. Modified from Nguyen et al. (2009).
IV. PROTEOME-WIDE CHARACTERIZATION OF NUCLEOTIDE–PROTEIN INTERACTIONS AND DISCOVERY OF NOVEL NUCLEOTIDE-BINDING TARGETS
Apart from protein kinases and GTP-binding proteins, a variety of proteins or protein complexes associate with different types of small nucleotide ligands to modulate cellular functions. For example, many ATP-binding proteins without kinase activity such as ATP-binding cassette transporters and chaperones, are involved in various pivotal cellular processes including cell signaling, proliferation, differentiation, and apoptosis (Chene, 2002). Nicotinamide adenine dinucleotide (NAD), as well as its phosphorylated and reduced forms, NADP+, NADH and NADPH, interacts with multiple enzymes to play critical roles in cellular metabolism, energy production, and cellular pathways like glycolysis and photosynthesis (Ansari & Raghava, 2010). In addition, cyclic nucleotide monophosphate like cAMP and cGMP are important intracellular secondary messenger molecules produced in response to hormone action. Protein kinase A subunits, cGMP-dependent protein kinases, as well as Rap guanine nucleotide exchange factors interact with cAMP and cGMP to regulate signaling cascades, whereas a number of ion channels can also bind to cAMP to modulate their activity (Luo et al., 2009). On the other hand, nucleotides are susceptible to damage from exposure to various genotoxic agents, including reactive oxygen species formed from normal metabolism or from exposure to ionizing radiation and environmental chemicals (Tsuchimoto et al., 2010). For example, 8-oxo-7,8-dihydroguanosine triphosphate (8-oxoGTP) could be produced at appreciable levels in the cytoplasm (Yoon et al., 2005). Some damaged nucleotides may compromise the flow of genetic information through their incorporation into RNA or DNA (Cheng et al., 1992; Kamiya & Kasai, 1995), whereas others may perturb cellular functions by binding to nucleotide-binding proteins (Yoon et al., 2005).
Despite the importance of nucleotide-binding proteins in almost every cellular process, much remains to be learned about protein–nucleotide interactions in the human proteome. For example, although a large number of known ATP-, NAD-, cAMP-, and cGMP-binding proteins are documented in gene ontology (GO) database, numerous proteins with available sequence information remain unannotated with respect to their nucleotide-binding affinity (Zhang et al., 2012). Additionally, there is no database available for proteins that can bind to damaged nucleotides. Furthermore, experimental characterization of nucleotide–protein interaction often relies on radioactivity-based ultrafiltration assay (Ormö & Sjöberg, 1990) or fluorescence-based binding assay (Guarnieri, Blagg, & Zhao, 2011). These traditional methods are usually costly and time-consuming because they require the use of purified proteins, which prevent high-throughput studies at the whole proteome level. Therefore, new methods for systematic characterizations of protein–nucleotide interactions at a global proteome scale are invaluable for understanding better the regulatory mechanisms of nucleotide-related protein functions.
Similar as kinase enrichment, strategies of affinity chromatography and ABPP can be used to facilitate the study of nucleotide–protein interaction and discovery of novel nucleo-tide-binding proteins. Various ribonucleotide (NTP) and 2′-deoxyribonucleotide (dNTP) ligands, such as CTP, UTP, dCTP, dUTP, dTTP, cAMP, and cGMP, have been immobilized onto agarose beads via flexible linkers (Cukor & Nowak, 1982; Scholten et al., 2006) and most of them are commercially available, which allows for convenient and rapid pull-down of nucleotide-binding proteins and discovery of novel nucleotide-binding targets by mass spectrometry. One drawback for this affinity chromatography strategy is that the pull-down fractions of protein mixture are often dominated by highly abundant proteins with low binding affinity to AMP/ADP/ATP, GMP/GDP/GTP, or DNA/RNA. Therefore, a sequential elution protocol was generally required to remove contaminated proteins. One successful example was the study of cAMP/cGMP-interacting proteome (interactome) (Scholten et al., 2006). In that study, after loading the whole cell lysates onto cAMP/cGMP affinity column, washing solutions containing ADP, GDP, cGMP, and/or cAMP were used to sequentially elute highly abundant ADP-, GDP-, and DNA-binding proteins from the column. Only the proteins remaining on the beads after the sequential elution steps were considered as strong cAMP/cGMP-binding proteins and were finally eluted by boiling with SDS-loading buffer for MS analysis. With the use of this protocol, the authors successfully validated the binding affinity of many known cAMP/cGMP-binding proteins such as PKA and PKG, several phosphodiesterases and several indirect cAMP/cGMP binders (e.g., A-kinase anchoring proteins). Similarly, this affinity chromatography strategy coupled with MS analysis has also been applied for comprehensive screening of damaged nucleotide-binding proteins from whole cell lysates (Tsuchimoto et al., 2010). In this screening system, various damaged nucleotides were immobilized onto affinity resins for the purification of damaged nucleotide-binding proteins, and the purified proteins were further identified by MS. For instance, these authors performed comprehensive screenings for inosine triphosphate (ITP)-binding proteins from mouse and human cell extracts, and nucleoside diphosphate linked moiety X-type motif 16 (NUDT16) protein was identified as ITP-binding enzymes. This pioneering report represents the first work for proteome-wide damaged nucleotide-binding protein screening, and this screening strategy can, in principle, be applied to study any kinds of damaged nucleotide-binding proteins. On the other hand, an ABPP approach was developed to profile the cAMP-interacting proteome (Luo et al., 2009). In this report, these authors designed, synthesized, and optimized a series of cAMP reactive affinity probes, or capture compounds. The cAMP capture compounds are trifunctional molecules that are comprised of a selectivity group (cAMP), photoactivatable cross-linking group, and a sorting group, for example, biotin. Therefore, once binding to the cAMP-binding proteins, the capture compound enables the covalent linkage to the target proteins by a photoactivatable functional group, followed by isolation of capture compound-protein conjugate through bio-tin-avidin interactions. Coupled with LC-MS, this ABPP approach was applied to study the cAMP-binding proteins from lysates of Escherichia coli, HepG2 cells, and subcellular fractions of mammalian brain, which showed better sensitivity than cAMP affinity chromatography approach (Scholten et al., 2006). Especially, the results suggest this cAMP ABPP strategy is uniquely efficient and sensitive for identification and profiling of cAMP-binding membrane proteins including multiple ion channels.
Although strategies discussed above achieved a great success in determining nucleotide–protein interaction, they provide limited information about the nucleotide-binding site, or “nucleotide-interacting residues,” which is another aspect of significant interest in nucleotide–protein interaction studies. For instance, the ATP binding sites, where ATP is captured and hydrolyzed to ADP, are particularly important because they are responsible for changing the conformation and/or modulating the catalytic activity of proteins (Chene, 2002). Additionally, many anti-cancer drugs target ATP-binding sites in ATPases and kinases (Chene, 2002). Although some bioinformatic tools were developed to identify nucleotide-interacting residues in proteins (Ansari & Raghava, 2010; Zhang et al., 2012), these tools still suffer from high false-positive rates and require experimental verification. To overcome these difficulties, we recently developed a quantitative affinity profiling strategy, encompassing the use of low and high concentrations of desthiobiotin-conjugated acyl ATP probes, to comprehensively characterize ATP-protein interactions at the entire proteome scale (Xiao et al., 2013b). As reported previously (Xiao et al., 2013a), the relatively high reactivity of the acyl ATP probes leads to their conjugation with not only the lysine residue(s) located at the nucleotide binding site, but also other lysine residues through non-specific electrostatic interactions. To differentiate specific from non-specific labelings, we developed a novel strategy to unambiguously characterize ATP-binding affinities of proteins at the entire proteome scale. In this vein, a previous study employed reactivity profiling for the proteome-wide discovery of proteins containing functional cysteines by comparing the extent of cysteine alkylation under different probe concentrations (Weerapana et al., 2010). Viewing that probe binding renders the acyl phosphate moiety conducive for coupling with the lysine residue at the ATP binding site and augments greatly the rate for amide bond formation (Li & Liu, 2004), lysine residues participating in ATP binding and those that are not would exhibit distinct labeling behaviors upon labeling with low and high concentrations of the ATP affinity probe. At a low probe concentration, the lysine residue at the ATP-binding site is completely labeled, whereas the lysines not involved in ATP binding are only partially labeled because limited amount of the labeling reagent reacts preferentially with the lysine at the ATP-binding site. At a high probe concentration, the ATP-binding lysine is still labeled to completion; however, non-ATP binding lysine is labeled to a much greater extent than that at low probe concentration (Fig. 10A). Based on the above analysis, we devised an affinity profiling strategy by allowing a low (10 µM) concentration of the light and heavy ATP probes and a high (100 µM) concentration of the heavy and light ATP probes, respectively, to react separately with the same amount of cell lysates. As a result, peak intensity ratios of light and heavy desthiobiotin-labeled peptides can be employed to derive ATP-binding affinity ratio, RATP10/1, which reflects the relative binding affinities of ATP towards specific lysine residues in individual proteins. Therefore, specific ATP-binding lysine will yield an RATP10/1 close to 1 since similar amount of ATP-binding lysine will be labeled regardless of the probe concentration. By contrast, non-specifically labeled lysine will manifest a concentration-dependent increase in ATP probe labeling, which results in an RATP10/1 ≫ 1 (Fig. 10A).
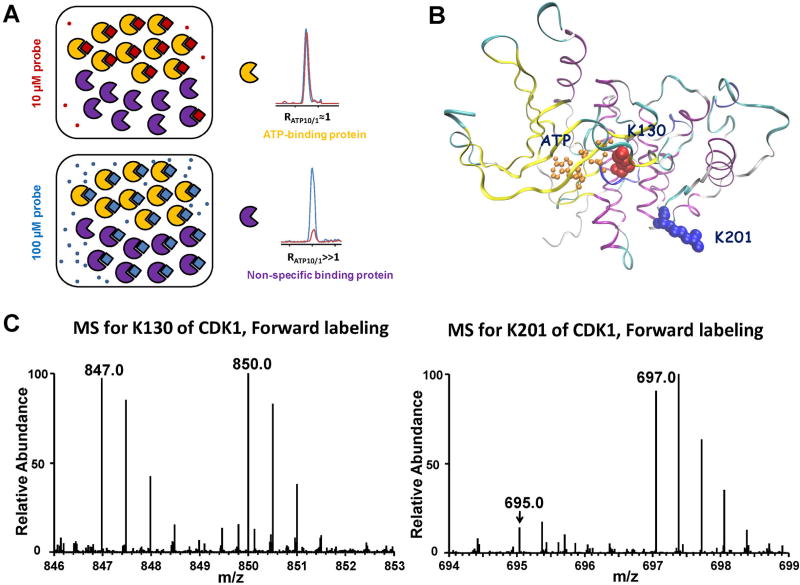
FIGURE 10.
A: A Cartoon representation showing expected labeling behavior of proteins with ATP-binding affinity (yellow) or without ATP-binding affinity (purple) treated with a low (10 µM) and high (100 µM) concentrations of the ATP-probe. At the low probe concentration, the ATP-binding protein labels to completion, but the non-specific binding protein is only partially labeled. At high probe concentration, the ATP-binding protein is still labeled to completion, whereas the non-specific binding protein shows much greater labeling. Modified from Weerapana et al. (2010). B: Crystal structure of CDK1 homologue, CDK2, bound with ATP displays the structural relationship between K130, K201, and ATP-binding sites on CDK1. C: Peptide DLK130#PQNLLIDDK with a low RATP10/1 ratio from cyclin-dependent kinase 1; Peptide K201#PLFHGDSEIDQLFR with a high RATP10/1 ratio from cyclin-dependent kinase 1. “#” Denotes the desthiobiotin-labeling site.
We demonstrated that this quantitative nucleotide-binding affinity profiling strategy allowed for site-specific determination of relative ATP-binding affinities of different binding sites in proteins by monitoring RATP10/1, thereby minimizing false-positive identification of ATP-binding sites. For instance, two lysine residues, K130 and K201, from cyclin-dependent kinase 1 were labeled in our quantitative profiling experiment; however, K201 exhibited a much larger RATP10/1 than K130, indicating that K130 is the true ATP-binding lysine and the labeling of K201 originates from non-specific binding (Fig. 10C). The crystal structure of Cdk1 homolog, Cdk2 (PDB entry: 1HCK), reveals the close proximity of K130 to the γ-phosphate group of ATP (Fig. 10B). Additionally, we demonstrated that this novel quantitative ATP-affinity profiling strategy is particularly useful for unveiling novel ATP-binding proteins and previously unrecognized nucleotide-binding sites in ATP-binding proteins. For instance, our profiling results led to the discovery of human proliferating cell nuclear antigen (PCNA) as a new ATP-binding protein and the identification of lysine residue that is involved in ATP binding (Xiao et al., 2013b).
By applying a similar nucleotide-binding affinity profiling strategy, we characterized comprehensively the binding property for the entire human proteome toward a modified nucleotide, 6-thioguanine triphosphate (SGTP). We identified 165 SGTP-binding proteins, among which GTPases and multiple hetero-trimeric G proteins display strong binding affinity toward SGTP. We also demonstrated that SGTP binds to several cyclin-dependent kinases (CDKs), which may perturb the CDK-mediated phosphorylation and cell cycle progression (Vassilev et al., 2006; Koledova et al., 2010). It can be envisaged that this nucleotide-binding affinity profiling strategy can be generally applicable for the future characterization of the interaction of other modified nucleotides with the global proteome. Such studies should also result in the discovery of specific proteins that can bind to these damaged nucleotides and how these proteins recognize damaged nucleotides versus their endogenous undamaged counterparts.
V. SUMMARY AND PERSPECTIVES
Nucleotide-binding proteins play pivotal roles in many cellular processes, particularly in cell signaling; however, global study of the nucleotide-binding proteins can hardly be achieved with the use of the conventional approaches, rendering the current knowledge of nucleotide–protein interactions far from complete. MS has become increasingly the method of choice for analysis of complex protein samples due to its high specificity, accuracy and throughput (Yates, Ruse, & Nakorchevsky, 2009). Recent developments in mass spectrometry instrumentation (Olsen et al., 2009), sample preparation methods (Wisniewski et al., 2009), and bioinformatics tools (Cox & Mann, 2008) have enabled proteome-wide characterizations of nucleotide–protein interactions. However, currently targeted study of sub-proteome of nucleotide-binding proteins, especially protein kinases and GTP-binding proteins, remains challenging owing to the high degrees of complexity of cellular proteomes and the relatively low abundances of many of these signaling proteins (Aebersold & Goodlett, 2001). Therefore, further technological advances in investigating nucleotide–protein interactions are highly desirable.
Firstly, it is widely recognized that pre-enrichment steps are necessary for facilitating MS-based proteomics study of nucleotide-binding proteins. Affinity chromatography and activity-based protein profiling with the use of chemically reactive affinity probes have been demonstrated to be the two most promising enrichment approaches for nucleotide-binding proteins, especially for the kinome. Among them, kinobeads and ATP-acyl phosphate reactive affinity probes exhibited great potential in profiling the expression and activity of the entire kinome, as well as in the discovery of novel targets for kinase inhibitors. While kinase enrichment with the use of kinobeads exhibited better selectivity than that with ATP-affinity reactive probes due to their distinct capture mechanisms, ATP-affinity probe holds its unique advantages in versatile applications by covalently modifying specific lysine residues within the conserved kinase sequences. In addition, the incorporation of an isotope-coded linker in ATP-affinity probes enables direct labeling of kinases to facilitate the accurate quantification of clinical samples. More importantly, the predictable biotin-modified lysine residue within the unique ATP-binding motif from captured kinase peptides allows for the confident identification and quantification of kinases with minimum false-positive rates.
Much remains to be improved for these enrichment platforms in terms of sensitivity and selectivity. Dozens of novel kinase inhibitors may serve as the potential candidates to be immobilized on the kinobeads to improve the coverage for kinome enrichment. For instance, a structure-guided bioinformatic analysis revealed that more than 200 kinases (approximately 40% of the protein kinome) have cysteine residues that can be covalently and irreversibly modified by novel irreversible kinase inhibitors (Zhang, Yang, & Gray, 2009; Garuti, Roberti, & Bottegoni, 2011; Liu et al., 2013; Sanderson, 2013). These irreversible kinase inhibitors, which possess different inhibitory mechanisms from current kinase inhibitors used for generating kinobeads, could serve as another type of ligands for kinobeads to expand the kinome coverage. In addition, affinity reactive probes based on widely used conjugation technique, such as click chemistry, have been developed for various classes of enzymes and drug targets (Adam, Sorensen, & Cravatt, 2002a,b; Barglow & Cravatt, 2007). It can be envisaged that reactive chemical affinity probes using kinase inhibitor as the affinity moiety may also be designed for kinase enrichment. Furthermore, anti-peptide or motif-specific antibodies have gained increasing interest (Kuhn et al., 2009). Sequential enrichment of targeted protein and then specific tryptic peptides from the protein by immunoaffinity purification with the use of anti-protein and anti-peptide antibodies from a clinical or biological sample prior to MS analysis was shown to increase the sensitivity of the assay by >104 (Neubert et al., 2013; Palandra et al., 2013). This is necessary and sufficient to render the assay sensitive enough for measurements of clinical or biological specimens. Nucleotide-binding proteins, especially protein kinases, ATPases, and GTPases often share a conserved sequence motif(s) as discussed above; therefore, orthogonal platforms encompassing enrichment at both the protein (such as kinobeads or reactive affinity probes) and peptide (using newly developed motif-specific antibodies) levels may greatly improve the selectivity and sensitivity of nucleotide detection workflows.
Most MS-based nucleotide-binding protein studies are often conducted in the data-dependent acquisition mode (Olsen et al., 2009). The inadequate sensitivity and reproducibility of these DDA detection strategies hamper their utility in biomarker discovery and clinical studies. Therefore, the MRM-based targeted MS analysis is more suitable for this multiplexed nucleotide-binding protein studies (Marx, 2013). The MRM-based targeted MS analysis permits rapid and continuous monitoring of specific ions of interest, which enhances the sensitivity for peptide detection by up to 100-fold relative to MS analysis in DDA-based discovery mode (Lange et al., 2008). More importantly, unlike shotgun proteomic technique, SRM analysis on triple quadrupole instruments yields a dynamic range of up to five orders of magnitude (Lange et al., 2008). Thus, the MRM-based targeted proteomic approach often affords much better reproducibility for quantification than shotgun proteomic method, rendering the MRM-based method particularly attractive in biological applications requiring repeated measurements of predefined set of proteins (Lange et al., 2008). Recently, affinity enrichment coupled with MRM has become increasingly employed for large-scale targeted analysis. For instance, targeted quantification of N-glycoproteins from 120 human plasma samples using MRM assay was recently reported (Hüttenhain et al., 2013). We also demonstrated that MRM coupled with reactive ATP-affinity probes can greatly improve the sensitivity and reproducibility for large-scale kinome detection (Xiao et al., 2014a). Moreover, other novel targeted MS analysis methods, including data-independent mass spectrometric acquisition (DIA) (Egertson et al., 2013) and sequential window acquisition of all theoretical spectra (SWATH) (Gillet et al., 2012; Rost et al., 2014), have been developed to achieve consistent and reproducible quantification. Due to the development of recombinant protein purification and peptide synthesis technologies, the MS/MS spectral libraries for the entire kinome or GTP-binding proteins, which serve as a resource for MRM- or DIA-based proteomic workflows, can be readily obtained. Therefore, these targeted MS analysis technologies can be readily coupled to various kinase or GTPase enrichment strategies to further facilitate the detection of nucleotide-binding proteins. For example, kinobeads exhibited better specificity than acyl-phosphate ATP probes for kinome detection, and the coupling of kinobeads with targeted MRM or DIA analysis may further increase its sensitivity and reliability.
In summary, the coupling of various enrichment platforms with targeted MS acquisition strategies such as MRM or DIA has and will continue to improve the sensitivity, coverage and reliability of nucleotide-binding protein analysis by MS.
References
- Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol. 2002a;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- Adam GC, Sorensen EJ, Cravatt BF. Trifunctional chemical probes for the consolidated detection and identification of enzyme activities from complex proteomes. Mol Cell Proteomics. 2002b;1:828–835. doi: 10.1074/mcp.t200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101:269–296. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- Ansari HR, Raghava GP. Identification of NAD interacting residues in proteins. BMC Bioinform. 2010;11:160. doi: 10.1186/1471-2105-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansong C, Ortega C, Payne SH, Haft DH, Chauvignè-Hines LM, Lewis MP, Ollodart AR, Purvine SO, Shukla AK, Fortuin S, Smith RD, Adkins JN, Grundner C, Wright AT. Identification of widespread adenosine nucleotide binding in Mycobacterium tuberculosis. Chem Biol. 2013;20:123–133. doi: 10.1016/j.chembiol.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007a;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: A critical review. Anal Bioanal Chem. 2007b;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Barglow KT, Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- Bartlett S, Beddard GS, Jackson RM, Kayser V, Kilner C, Leach A, Nelson A, Oledzki PR, Parker P, Reid GD, Warriner SL. Comparison of the ATP binding sites of protein kinases using conformationally diverse bisindolylmaleimides. JAm Chem Soc. 2005;127:11699–11708. doi: 10.1021/ja050576u. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Borisov OV, Goshe MB, Conrads TP, Rakov VS, Veenstra TD, Smith RD. Low-energy collision-induced dissociation fragmentation analysis of cysteinyl-modified peptides. Anal Chem. 2002;74:2284–2292. doi: 10.1021/ac010974p. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene P. ATPases as drug targets: Learning from their structure. Nat Rev Drug Discov. 2002;1:665–673. doi: 10.1038/nrd894. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—T and A—C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cooper MJ, Cox NJ, Zimmerman EI, Dewar BJ, Duncan JS, Whittle MC, Nguyen TA, Jones LS, Ghose Roy S, Smalley DM, Kuan PF, Richards KL, Christopherson RI, Jin J, Frye SV, Johnson GL, Baldwin AS, Graves LM. Application of multiplexed kinase inhibitor beads to study kinome adaptations in drug-resistant leukemia. PLoS ONE. 2013;8:e66755. doi: 10.1371/journal.pone.0066755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. Ras family signaling: Therapeutic targeting. Cancer Biol Ther. 2002;1:599–606. doi: 10.4161/cbt.306. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Cukor G, Nowak NA. Affinity chromatography purification of Pseudomonas aeruginosa exotoxin A on specifically linked NAD agarose. J Bacteriol. 1982;149:1162–1165. doi: 10.1128/jb.149.3.1162-1165.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phospho-proteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- Dever TE, Glynias MJ, Merrick WC. GTP-binding domain: Three consensus sequence elements with distinct spacing. Proc Natl Acad Sci USA. 1987;84:1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyrup AT, Krishnan S, Cockburn BN, Schwartz NB. Deletion and site-directed mutagenesis of the ATP-binding motif (P-loop) in the bifunctional murine Atp-sulfurylase/adenosine 5-phosphosulfate kinase enzyme. J Biol Chem. 1998;273:9450–9456. doi: 10.1074/jbc.273.16.9450. [DOI] [PubMed] [Google Scholar]
- Dong X, Xiao Y, Jiang X, Wang Y. Quantitative proteomic analysis revealed lovastatin-induced perturbation of cellular pathways in HL-60 cells. J Proteome Res. 2011;10:5463–5471. doi: 10.1021/pr200718p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan P-F, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham WH, Mullin M, Gingras A-C. Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, Wu CC, MacCoss MJ. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, Rinner O. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustace AJ, Dowling P, Henry M, Doolan P, Meleady P, Clynes M, Crown J, O’Donovan N. 2D–DIGE analysis of phospho-enriched fractions from dasatinib-treated melanoma cell lines. J Proteomics. 2011;74:490–501. doi: 10.1016/j.jprot.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias J-M, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Garuti L, Roberti M, Bottegoni G. Irreversible protein kinase inhibitors. Curr Med Chem. 2011;18:2981–2994. doi: 10.2174/092986711796391705. [DOI] [PubMed] [Google Scholar]
- George Cisar EA, Nguyen N, Rosen H. A GTP affinity probe for proteomics highlights flexibility in purine nucleotide selectivity. J Am Chem Soc. 2013;135:4676–4679. doi: 10.1021/ja400839e. [DOI] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Kwiek JJ, Fadden P, Ray R, Hardeman K, Coley AM, Foley M, Haystead TAJ. Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol Pharmacol. 2002;62:1364–1372. doi: 10.1124/mol.62.6.1364. [DOI] [PubMed] [Google Scholar]
- Guarnieri MT, Blagg BS, Zhao R. A high-throughput TNP-ATP displacement assay for screening inhibitors of ATP-binding in bacterial histidine kinases. Assay Drug Dev Technol. 2011;9:174–183. doi: 10.1089/adt.2010.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Xiao Y, Wang Y. Monomethylarsonous acid inhibited endogenous cholesterol biosynthesis in human skin fibroblasts. Toxicol Appl Pharmacol. 2014;277:21–29. doi: 10.1016/j.taap.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanoulle X, Van Damme J, Staes A, Martens L, Goethals M, Vandekerck-hove J, Gevaert K. A new functional, chemical proteomics technology to identify purine nucleotide binding sites in complex proteomes. J Protoeme Res. 2006;5:3438–3445. doi: 10.1021/pr060313e. [DOI] [PubMed] [Google Scholar]
- Horscroft NJ, Roy P. NTP binding and phosphohydrolase activity associated with purified bluetongue virus non-structural protein NS2. J Gen Virol. 2000;81:1961–1965. doi: 10.1099/0022-1317-81-8-1961. [DOI] [PubMed] [Google Scholar]
- Hüttenhain R, Surinova S, Ossola R, Sun Z, Campbell D, Cerciello F, Schiess R, Bausch-Fluck D, Rosenberger G, Chen J, Rinner O, Kusebauch U, Hajdúch M, Moritz RL, Wollscheid B, Aebersold R. N-Glycoprotein SRMAtlas: A resource of mass spectrometric assays for N-glycosites enabling consistent and multiplexed protein quantification for clinical applications. Mol Cell Proteomics. 2013;12:1005–1016. doi: 10.1074/mcp.O112.026617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Heazlewood JL, Millar AH. Analysis of the soluble ATP-binding proteome of plant mitochondria identifies new proteins and nucleotide triphosphate interactions within the matrix. J Proteome Res. 2006;5:3459–3469. doi: 10.1021/pr060403j. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, Garraway LA. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN, Noble MEM, Owen DJ. Active and inactive protein kinases: Structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Kamiya H, Kasai H. Formation of 2-hydroxydeoxyadenosine triphos-phate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases: Steady-state kinetics of the incorporation. J Biol Chem. 1995;270:19446–19450. doi: 10.1074/jbc.270.33.19446. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Masuda S, Tomohiro T, Hatanaka Y. A simple and efficient photoaffinity method for proteomics of GTP-binding proteins. Chem-BioChem. 2007;8:595–598. doi: 10.1002/cbic.200600527. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci USA. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koledova Z, Kafkova LR, Calabkova L, Krystof V, Dolezel P, Divoky V. Cdk2 inhibition prolongs G1 phase progression in mouse embryonic stem cells. Stem Cells Dev. 2010;19:181–194. doi: 10.1089/scd.2009.0065. [DOI] [PubMed] [Google Scholar]
- Kruse U, Pallasch CP, Bantscheff M, Eberhard D, Frenzel L, Ghidelli S, Maier SK, Werner T, Wendtner CM, Drewes G. Chemoproteo-mics-based kinome profiling and target deconvolution of clinical multi-kinase inhibitors in primary chronic lymphocytic leukemia cells. Leukemia. 2011;25:89–100. doi: 10.1038/leu.2010.233. [DOI] [PubMed] [Google Scholar]
- Ku X, Heinzlmeir S, Helm D, Médard G, Kuster B. New affinity probe targeting VEGF receptors for kinase inhibitor selectivity profiling by chemical proteomics. J Protoeme Res. 2014a;13:2445–2452. doi: 10.1021/pr401247t. [DOI] [PubMed] [Google Scholar]
- Ku X, Heinzlmeir S, Liu X, Médard G, Kuster B. A new chemical probe for quantitative proteomic profiling of fibroblast growth factor receptor and its inhibitors. J Proteomics. 2014b;96:44–55. doi: 10.1016/j.jprot.2013.10.031. [DOI] [PubMed] [Google Scholar]
- Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, Sabatine MS, Gerszten RE, Carr SA. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009;55:1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeer S, Zörgiebel C, Ruprecht B, Kohl K, Kuster B. Comparing immobilized kinase inhibitors and covalent ATP probes for proteomic profiling of kinase expression and drug selectivity. J Protoeme Res. 2013;12:1723–1731. doi: 10.1021/pr301073j. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu DR. DNA-templated organic synthesis: Nature’s strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem Int Ed. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage Sara J, Jones Lyn H, Gray Nathanael S. Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo Tejedor M, Mizuno H, Tsuyama N, Harada T, Masujima T. In situ molecular analysis of plant tissues by live single-cell mass spectrometry. Anal Chem. 2011;84:5221–5228. doi: 10.1021/ac202447t. [DOI] [PubMed] [Google Scholar]
- Luo Y, Blex C, Baessler O, Glinski M, Dreger M, Sefkow M, Köster H. The cAMP capture compound mass spectrometry as a novel tool for targeting cAMP-binding proteins: From protein kinase A to potassium/ sodium hyperpolarization-activated cyclic nucleotide-gated channels. Mol Cell Proteomics. 2009;8:2843–2856. doi: 10.1074/mcp.M900110-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Marx V. Targeted proteomics. Nat Methods. 2013;10:19–22. doi: 10.1038/nmeth.2285. [DOI] [PubMed] [Google Scholar]
- McAllister FE, Niepel M, Haas W, Huttlin E, Sorger PK, Gygi SP. Mass spectrometry based method to increase throughput for kinome analyses using ATP probes. Anal. Chem. 2013;85:4666–4674. doi: 10.1021/ac303478g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert H, Muirhead D, Kabir M, Grace C, Cleton A, Arends R. Sequential protein and peptide immunoaffinity capture for mass spectrometry-based quantification of total human beta-nerve growth factor. Anal Chem. 2013;85:1719–1726. doi: 10.1021/ac303031q. [DOI] [PubMed] [Google Scholar]
- Newton AC. Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- Nguyen UT, Guo Z, Delon C, Wu Y, Deraeve C, Franzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, Wolters D, Alexandrov K. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases: Controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat Rev Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormö M, Sjöberg B-M. An ultrafiltration assay for nucleotide binding to ribonucleotide reductase. Anal Biochem. 1990;189:138–141. doi: 10.1016/0003-2697(90)90059-i. [DOI] [PubMed] [Google Scholar]
- Pachl F, Plattner P, Ruprecht B, Me´dard G, Sewald N, Kuster B. Characterization of a chemical affinity probe targeting Akt kinases. J Protoeme Res. 2013;12:3792–3800. doi: 10.1021/pr400455j. [DOI] [PubMed] [Google Scholar]
- Palandra J, Finelli A, Zhu M, Masferrer J, Neubert H. Highly specific and sensitive measurements of human and monkey interleukin 21 using sequential protein and tryptic peptide immunoaffinity LC-MS/MS. Anal Chem. 2013;85:5522–5529. doi: 10.1021/ac4006765. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee J-D, Gray NS, Kozarich JW. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2006;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R. Selected reaction monitoring-based proteo-mics: Workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- Preininger AM, Hamm HE. G protein signaling: Insights from new structures. Sci Signal. 2004;2004:re3. doi: 10.1126/stke.2182004re3. [DOI] [PubMed] [Google Scholar]
- Pulford K, Lamant L, Morris SW, Butler LH, Wood KM, Stroud D, Delsol G, Mason DY. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89:1394–1404. [PubMed] [Google Scholar]
- Qiu H, Wang Y. Probing adenosine nucleotide-binding proteins with an affinity-labeled nucleotide probe and mass spectrometry. Anal Chem. 2007;79:5547–5556. doi: 10.1021/ac0622375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: Role of ATP binding site in suppression of caspace-1 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- Rosenblum JS, Nomanbhoy TK, Kozarich JW. Functional interrogation of kinases and other nucleotide-binding proteins. FEBS Lett. 2013;587:1870–1877. doi: 10.1016/j.febslet.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein auantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Rost HL, Rosenberger G, Navarro P, Gillet L, Miladinovic SM, Schubert OT, Wolski W, Collins BC, Malmstrom J, Malmstrom L, Aebersold R. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- Sanderson K. Irreversible kinase inhibitors gain traction. Nat Rev Drug Discov. 2013;12:649–651. doi: 10.1038/nrd4103. [DOI] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop—A common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Scholten A, Poh MK, van Veen TA, van Breukelen B, Vos MA, Heck AJ. Analysis of the cGMP/cAMP interactome using a chemical proteomics approach in mammalian heart tissue validates sphingosine kinase type 1-interacting protein as a genuine and highly abundant AKAP. J Proteome Res. 2006;5:1435–1447. doi: 10.1021/pr0600529. [DOI] [PubMed] [Google Scholar]
- Shugar D. The NTP phosphate donor in kinase reactions: Is ATP a monopolist? Acta Biochim Pol. 1996;43:9–23. [PubMed] [Google Scholar]
- Sioud S, Genestie B, Jahouh F, Martin P, Banoub J. Gasphase fragmentation study of biotin reagents using electrospray ionization tandem mass spectrometry on a quadrupole orthogonal time-of-flight hybrid instrument. Rapid Commun Mass Spectrom. 2009;23:1941–1956. doi: 10.1002/rcm.4091. [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Taylor P, Nielsen PA, Trelle MB, Andersen MB, Vorm O, Moran MF, Kislinger T. Automated 2D peptide separation on a 1D Nano-LC-MS system. J Proteome Res. 2009;8:1610–1616. doi: 10.1021/pr800986c. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto D, Iyama T, Nonaka M, Abolhassani N, Ohta E, Sakumi K, Nakabeppu Y. A comprehensive screening system for damaged nucleotide-binding proteins. Mutat Res. 2010;703:37–42. doi: 10.1016/j.mrgentox.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Tsuyama N, Mizuno H, Tokunaga E, Masujima T. Live single-cell molecular analysis by video-mass spectrometry. Anal Sci. 2008;24:559–561. doi: 10.2116/analsci.24.559. [DOI] [PubMed] [Google Scholar]
- van Corven EJ, Groenink A, Jalink K, Eichholtz T, Moolenaar WH. Lysophosphatidate-induced cell proliferation: Identification and dissection of signaling pathways mediated by G proteins. Cell. 1989;59:45–54. doi: 10.1016/0092-8674(89)90868-4. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamor JG, Kaschani F, Colby T, Oeljeklaus J, Zhao D, Kaiser M, Patricelli MP, van der Hoorn RAL. Profiling protein kinases and other ATP binding proteins in arabidopsis using Acyl-ATP Probes. Mol Cell Proteomics. 2013;12:2481–2496. doi: 10.1074/mcp.M112.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, Smith RD. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark B, Johnsson A, Paulsson Y, Betsholtz C, Heldin CH, Herlyn M, Rodeck U, Koprowski H. Human melanoma cell lines of primary and metastatic origin express the genes encoding the chains of platelet-derived growth factor (PDGF) and produce a PDGF-like growth factor. Proc Natl Acad Sci USA. 1986;83:7197–7200. doi: 10.1073/pnas.83.19.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Wolfe LM, Veeraraghavan U, Idicula-Thomas S, Schuürer S, Wennerberg K, Reynolds R, Besra GS, Dobos KM. A chemical proteomics approach to profiling the ATP-binding proteome of Mycobacterium tuberculosis. Mol Cell Proteomics. 2013;12:1644–1660. doi: 10.1074/mcp.M112.025635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Guo L, Jiang X, Wang Y. Proteome-wide discovery and characterizations of nucleotide-binding proteins with affinity-labeled chemical probes. Anal Chem. 2013a;85:3198–3206. doi: 10.1021/ac303383c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Guo L, Wang Y. Isotope-coded ATP probe for quantitative affinity profiling of ATP-binding proteins. Anal Chem. 2013b;85:7478–7486. doi: 10.1021/ac401415z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Guo L, Wang Y. A targeted quantitative proteomics strategy for global kinome profiling of cancer cells and tissues. Mol Cell Proteomics. 2014a;13:1065–1075. doi: 10.1074/mcp.M113.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Ji D, Guo L, Wang Y. Comprehensive characterization of SGTP-binding proteins by orthogonal quantitative SGTP-affinity profiling and SGTP/GTP competition assays. Anal Chem. 2014b;86:4550–4558. doi: 10.1021/ac500588q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu K, Fujihara M, Tachibana M, Katayama S, Takahashi A, Hara E, Imai H, Shinkai Y, Yamashita Jun K. Protein kinase A determines timing of early differentiation through epigenetic regulation with G9a. Cell Stem Cell. 2012;10:759–770. doi: 10.1016/j.stem.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrome-try: Approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- Yoon S-H, Hyun J-W, Choi J, Choi E-Y, Kim H-J, Lee S-J, Chung M-H. In vitro evidence for the recognition of 8-oxoGTP by Ras, a small GTP-binding protein. Biochem Biophys Res Commun. 2005;327:342–348. doi: 10.1016/j.bbrc.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Zhang L, Holmes IP, Hochgraäfe F, Walker SR, Ali NA, Humphrey ES, Wu J, de Silva M, Kersten WJA, Connor T, Falk H, Allan L, Street IP, Bentley JD, Pilling PA, Monahan BJ, Peat TS, Daly RJ. Characterization of the novel broad-spectrum kinase inhibitor CTx-0294885 as an affinity reagent for mass spectrometry-based kinome profiling. J Protoeme Res. 2013;12:3104–3116. doi: 10.1021/pr3008495. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- Zhang YN, Yu DJ, Li SS, Fan YX, Huang Y, Shen HB. Predicting protein-ATP binding sites from primary sequence through fusing bi-profile sampling of multi-view features. BMC Bioinform. 2012;13:118. doi: 10.1186/1471-2105-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]