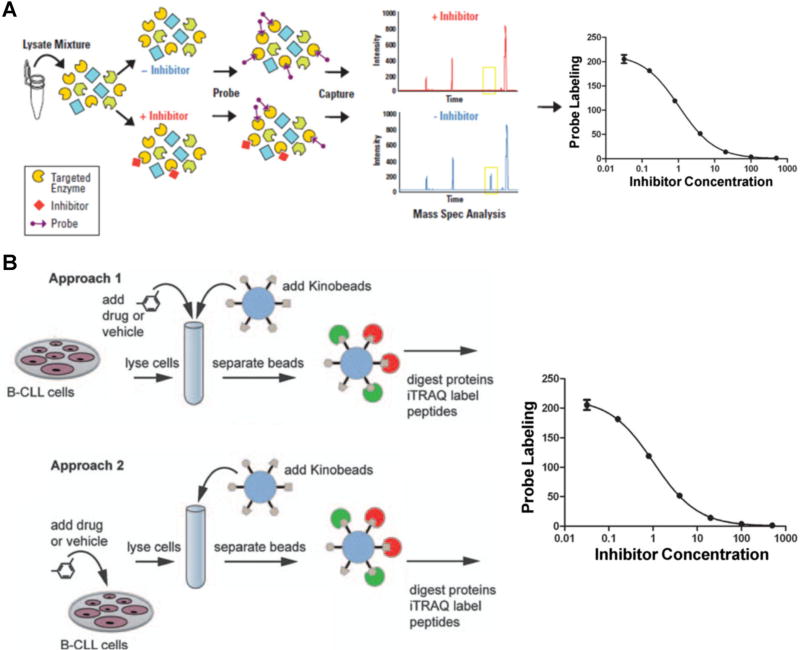
FIGURE 8.
Kinome profiling assays for the characterizations of the binding affinities and targets of kinase inhibitors. A: Workflow of a KiNativ experiment to determine the binding constants of kinase inhibitors. Whole proteome is treated with an inhibitor followed by addition of the reactive ATP affinity probe. After tryptic digestion and streptavidin enrichment, the probe-labeled kinase peptides with or without inhibitor treatment are quantified by LC-MS analysis. Modified from Patricelli et al. (2011). B: Workflow of kinobeads competition assay to determine drug targets. In the first approach, the drug of interest is added to the lysate at different concentrations followed by enrichment using the kinobeads, where the drug competes with the kinobeads for binding to protein kinases; in the second approach, live cells are kept in culture and treated with various concentrations of drug. Subsequently, cell lysates are generated and endogenous protein kinases are captured by kinobeads and analyzed by LC-MS. Modified from Kruse et al. (2011).