Figure 3. Sp110 induces ER stress responses in mouse macrophages.
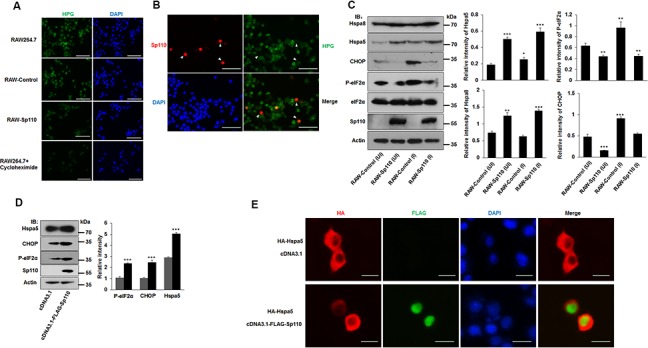
(A) The nascent protein synthesis in RAW264.7, RAW-Control and RAW-Sp110 cells were determined by HPG analysis. RAW264.7 cells treated with protein synthesis inhibitor cycloheximide (10 μg/ml, 12 h) were used as positive control. (B) RAW264.7 cells were transfected with the Sp110 expression plasmid for 24 h. Sp110 protein was detected by immunofluorescence staining, and protein synthesis was determined by HPG analysis. Nuclei were stained with DAPI. Scale bar = 50 μm. (C) Cell lysates prepared from the uninfected (UI) and H37Ra-infected ((MOI 5:1) RAW-Control and RAW-Sp110 cells (I) were analyzed by immunoblotting using antibodies against Hspa8, Hspa5, CHOP, P-eIF2α, eIF2α, Sp110, and Actin. The bands corresponding to Hspa8, Hspa5, CHOP, and P-eIF2α were quantified with ImageJ, and the intensity of each protein was normalized to the intensity of Actin. (D) RAW264.7 cells were cotransfected with pcDNA3.1 or pcDNA3.1-Flag-Sp110 for 48 h, the protein levels of Sp110, Hspa5, CHOP, and P-eIF2α were determined by immunoblotting. The bands corresponding to Hspa5, CHOP and P-eIF2α were quantified with ImageJ, and the intensity of each protein was normalized to the intensity of Actin. (E) RAW264.7 cells were cotransfected with pcDNA3.1 or pcDNA3.1-Flag-Sp110 and pCMV-HA-Hspa5 for 24 h, and then the subcellular localizations of Sp110 and Hspa5 were examined by immunofluorescence staining. Nuclei were stained with DAPI. Scale bar = 10 μm. Data are present as means ±SD of three independent experiments, * p< 0.05, **p< 0.01, and *** p< 0.001.
