Figure 1.
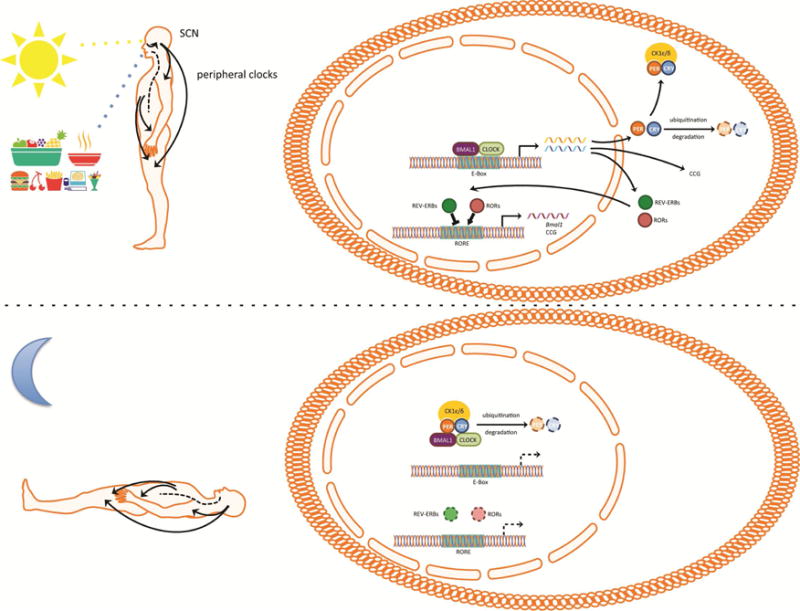
The mammalian circadian clock. Photosensitive melanopsin ganglion cells within the retina relay light information to the neurons of the suprachiasmatic nucleus (SCN). The neurons of the SCN fire with a rhythm of approximately 24 hours that is driven by autoregulatory transcriptional and translational feedback loops known as circadian clock. Circadian clocks exist in most cells and consist of transcriptional activators Brain Muscle Arnt-Like Protein 1 (BMAL1) and Clock Locomotor Output Kaput (CLOCK) that drive the expression of genes during the daytime. Among those genes are Period (Per) and Cryptochrome (Cry) that encode repressors of BMAL1 and CLOCK and Rors, Rev-erbs. PER and CRY form complexes with casein kinase 1 ε/δ (CK1ε/δ) that translocate to the nucleus during early night and repress the transcriptional activity of BMAL1 and CLOCK. Degradation of PER and CRY ends the repression on BMAL1 and CLOCK and allows the initiation of a new transcriptional cycle. Postranslational modifications of PER and CRY delay the transcriptional translational loop to a period of approximately 24 hours. The nuclear receptors RAR-related orphan receptor (ROR) (α, β, γ) and REV-ERB (α, β) translocate to the nucleus to activate and repress the expression of Bmal1. The circadian clocks of the SCN synchronize the clocks of the rest of the body to the same phase to generate 24 hour rhythms of physiology and behavior. Environmental cues such as feeding synchronize peripheral clocks but not the SCN clocks to the time of feeding.
