Figure 4.
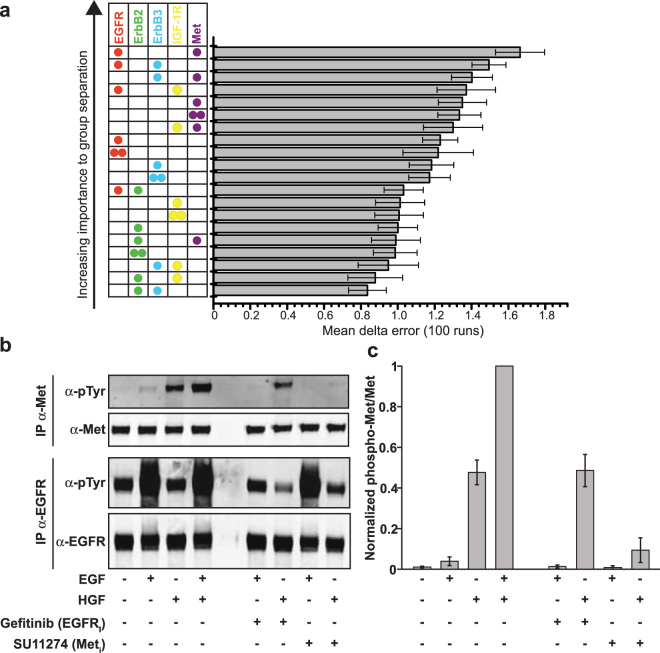
Random forest and biochemical analysis detect cross-stimulation of Met by EGFR following ligand exposure. (a) Random forest classification of non-linear combinations of features that reflect molecular interactions between receptors. Feature importance was calculated by repeatedly generating random forests from sub-samples of the data. (b) Biochemical analysis of Met-EGFR interaction. Serum-starved BT20 cells were pre-treated for 15 min with 1 µM Gefitinib for EGFR or SU11274 for Met or DMSO as a control and then stimulated for 5 min with EGF (which binds EGFR) and/or HGF (which binds Met) or medium as a control. Receptor immunoprecipitates were subjected to SDS-PAGE and immunoblots were probed with an anti-phospho-tyrosine antibody, anti-EGFR or anti-Met antibodies and respective secondary antibodies. The full length blots are presented in Supplementary Fig. 8. (c) Background-subtracted phospho-Met signals were corrected for total Met expression, normalized to the EGF/HGF-treated sample and plotted as mean with standard deviation derived from three biological replicates.