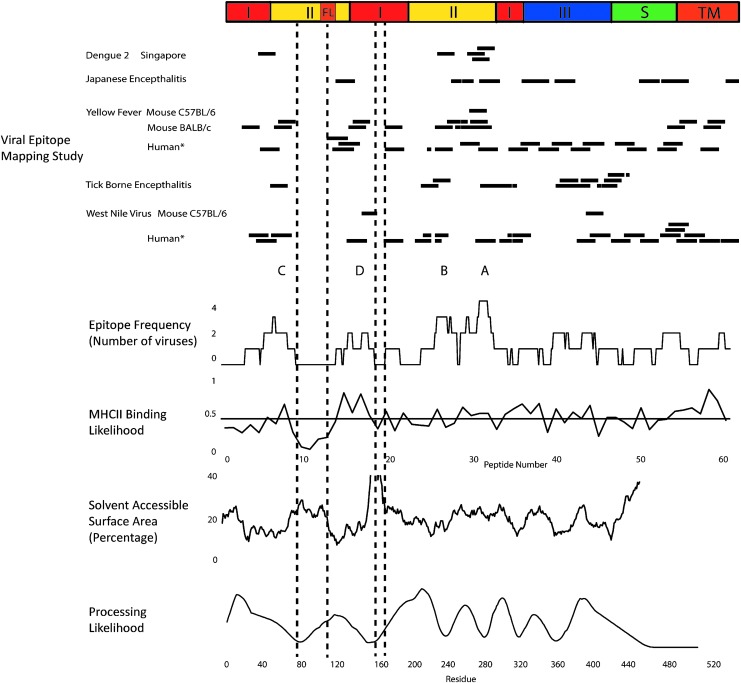
FIG. 2.
Epitope frequency aligned with predicted MHCII binding, solvent-accessible surface area, and processing likelihood. At top, a schematic diagram indicates structural domains identified in crystallographic structures. Horizontal bars indicate peptides identified as CD4+ epitopes or immunodominant epitope clusters (TBEV study) as they appear in the corresponding aligned E protein sequences. Epitope frequency was compiled as the number of viruses for which a given amino acid residue appears in an epitope or cluster, not including those identified by tetramer-guided mapping (*). MHCII binding likelihood indicates the average normalized prediction of binding affinity (“1-log50k”) for seven MHCII proteins (HLA-DRB1*0101, HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB1*0404, HLA-DRB1*0701, HLA-DRB1*1101, and HLA-DRB1*1501). The horizontal line indicates the mean for all sequence positions. Solvent-accessible area was calculated using MOLMOL with the crystal structure of the E protein from DFV2 (PDB: 1OK8) and then smoothed with a 15-residue averaging window. Processing likelihood was calculated for DFV2 using the algorithm previously described (34). HLA, human leukocyte antigen; MHCII, major histocompatibility class II; TBEV, tick-borne encephalitis virus; DFV2, dengue fever virus type 2.